Team:TUDelft/Temperature results
From 2008.igem.org
(→Setting up luciferase measurements) |
(→Setting up luciferase measurements) |
||
| Line 23: | Line 23: | ||
==Setting up luciferase measurements== | ==Setting up luciferase measurements== | ||
| + | |||
| + | ===Luminescence of luciferase can be measured in two strains=== | ||
The first two devices that were succesfully transformed were BBa_K115012 and BBa_K115034. First we wanted an indication whether it was possible to measure luciferase using these constructs, so an experiment was set up with these two strains. The exact protocol used can be found on the [https://2008.igem.org/TUDelft/19_September_2008 19th of September] of the lab notebook. In short, sonication and lysis buffer from the Promega Luciferase kit were used to lyse the cells and luciferase expression was normalized to OD<sub>600</sub> of the original sample (before lysis). Furthermore, we cells were grown with and without arabinose induction as an inducible promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_R0080 BBa_R0080]) was used. Figure 3 is a graphical representation of the amount of luciferase measured for the two strains the conditions depicted.<br/><br/> | The first two devices that were succesfully transformed were BBa_K115012 and BBa_K115034. First we wanted an indication whether it was possible to measure luciferase using these constructs, so an experiment was set up with these two strains. The exact protocol used can be found on the [https://2008.igem.org/TUDelft/19_September_2008 19th of September] of the lab notebook. In short, sonication and lysis buffer from the Promega Luciferase kit were used to lyse the cells and luciferase expression was normalized to OD<sub>600</sub> of the original sample (before lysis). Furthermore, we cells were grown with and without arabinose induction as an inducible promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_R0080 BBa_R0080]) was used. Figure 3 is a graphical representation of the amount of luciferase measured for the two strains the conditions depicted.<br/><br/> | ||
| Line 37: | Line 39: | ||
<br/> | <br/> | ||
Several conclusions can be drawn from figure 3, although it has to be noted that this result is only one measurement. Furthermore, room temperature was not controlled. It may have fluctuated between 20 and 27ºC. The first conclusion is that we are able to measure luciferase with these constructs. This means that all parts used work correctly, including our ribosome binding site. The second conclusion is that a differential amount of luminescence is measured between strains BBa_K115012 and BBa_K115034, although in these results we cannot see a difference that is temperature dependent. This can be seen when the bars in conditions 1 and 2 are compared, BBa_K115012 gives a larger difference in luminescence from room temperature to 37ºC than BBa_K115034. The second conclusion is that luminescence measured when lysis is performed with the provided buffer is much higher (compare bars of condition 4 to the rest). The condition measured is not optimal as room temperature is used rather than 37ºC, but even now luminescence is a multitude higher than any of the sonicated samples. The reason for this is not clear: it could be that the lysis buffer lyses cells more efficiently, alternatively it could be that sonication denaturates a large part of the luciferase in the cell. Another conclusion is that the arabinose promoter does not work; a similar amount of luminescence was measured in samples where arabinose was added as compared to non-induced samples (figure 3, compare 5 to 1, 2, and 3). | Several conclusions can be drawn from figure 3, although it has to be noted that this result is only one measurement. Furthermore, room temperature was not controlled. It may have fluctuated between 20 and 27ºC. The first conclusion is that we are able to measure luciferase with these constructs. This means that all parts used work correctly, including our ribosome binding site. The second conclusion is that a differential amount of luminescence is measured between strains BBa_K115012 and BBa_K115034, although in these results we cannot see a difference that is temperature dependent. This can be seen when the bars in conditions 1 and 2 are compared, BBa_K115012 gives a larger difference in luminescence from room temperature to 37ºC than BBa_K115034. The second conclusion is that luminescence measured when lysis is performed with the provided buffer is much higher (compare bars of condition 4 to the rest). The condition measured is not optimal as room temperature is used rather than 37ºC, but even now luminescence is a multitude higher than any of the sonicated samples. The reason for this is not clear: it could be that the lysis buffer lyses cells more efficiently, alternatively it could be that sonication denaturates a large part of the luciferase in the cell. Another conclusion is that the arabinose promoter does not work; a similar amount of luminescence was measured in samples where arabinose was added as compared to non-induced samples (figure 3, compare 5 to 1, 2, and 3). | ||
| + | |||
| + | ===Total protein content measurements in stead of OD measurements=== | ||
To compare the amount of luciferase expression measured, the total luciferase expression should be normalized. In the first experiment we normalized the expression to the OD<sub>600</sub> in the total culture. The OD<sub>600</sub> is an indication for the amount of cells present in the sample. However in this set up, luciferase does not leave the cell. This means cell lysis should be performed under conditions that leave the luciferase protein intact for measurements. This poses a challenge: because the efficiency of lysis is unknown and cannot be presumed constant, we will need to perform additional protein content measurements. | To compare the amount of luciferase expression measured, the total luciferase expression should be normalized. In the first experiment we normalized the expression to the OD<sub>600</sub> in the total culture. The OD<sub>600</sub> is an indication for the amount of cells present in the sample. However in this set up, luciferase does not leave the cell. This means cell lysis should be performed under conditions that leave the luciferase protein intact for measurements. This poses a challenge: because the efficiency of lysis is unknown and cannot be presumed constant, we will need to perform additional protein content measurements. | ||
{{Template:TUDelftiGEM2008_sidebar}} | {{Template:TUDelftiGEM2008_sidebar}} | ||
Revision as of 14:39, 28 October 2008
Contents |
Results
Assembly of BBa_K115012 and BBa_K115029 - BBa_K115036
The first challenge of this project was to assemble all the devices we wanted to measure. An overview of these devices can be found here. For the actual construction of all devices the 3A assembly strategy was chosen. A complete protocol of this strategy can be found on OpenWetWare. A schematic representation of this assembly method is depicted in figure 1A and 1B.
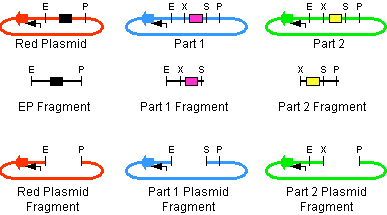
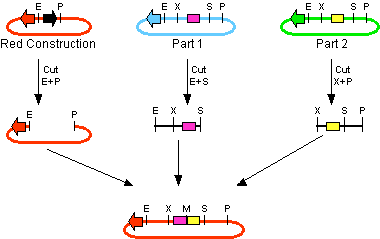
For the succesful use of this assembly, it is essential that the DB3.1 Eschirichia coli strain is not used as it tolerates the presence of the CcdB gene. Throughout the project we used commercial Top10 cells of Invitrogen that were made competent chemically. Furthermore, it is important to note that the XbaI and SpeI restriction enzymes generate compatible sticky ends. Assuming that we want to place two BioBrick parts behind each other in one plasmid, but these are present in seperate plasmids before assembly (as is the case in Figure 1A, parts are depicted in purple and yellow in the blue and green plasmid, respectively). 3A assembly can be conducted by taking an empty plasmid that has a different antibiotic resistence as the other two plasmids (red in figure 1A and B, CcdB is present). Three seperate restriction reactions should be started as shown in figure 1A. After cleaning up the restriction reactions they are mixed and ligated together. Parts can be ligated in several ways:
- The CcdB gene can be ligated back in the vector - If this happens, cells will not grow after transfection, as the E. coli strain used should not be CcdB gene tolerant.
- Other parts can be ligated back in the vector - Other plasmids should not have the same antibiotic resistence as the red plasmid, in other words, these are selected for.
- Ligation as depicted in figure 1B - This is the ligation we are looking for and it should yield colonies after transformation.
- Other combinations of backbones and/or parts - It is possible other combinations (e.g. the three backbones together) form a plasmid that yields colonies, these parts will be much larger as the ones formed in option 3. Colony PCR's are performed afterwards to screen for these.
In the end, the result on the gel of the colony PCR product is conclusive in terms of the succes of the 3A assembly. For all our parts at least three of the 3A assemblies (first promoter + ribosome binding site and luciferase + terminators, afterwards the result of these two together) gave our final product. An overview of the result on the gels can be found here. We succeeded in getting colonies that gave the correct band size on the gel for BBa_K115012 and BBa_K115029 - BBa_K115036.
Setting up luciferase measurements
Luminescence of luciferase can be measured in two strains
The first two devices that were succesfully transformed were BBa_K115012 and BBa_K115034. First we wanted an indication whether it was possible to measure luciferase using these constructs, so an experiment was set up with these two strains. The exact protocol used can be found on the 19th of September of the lab notebook. In short, sonication and lysis buffer from the Promega Luciferase kit were used to lyse the cells and luciferase expression was normalized to OD600 of the original sample (before lysis). Furthermore, we cells were grown with and without arabinose induction as an inducible promoter (BBa_R0080) was used. Figure 3 is a graphical representation of the amount of luciferase measured for the two strains the conditions depicted.
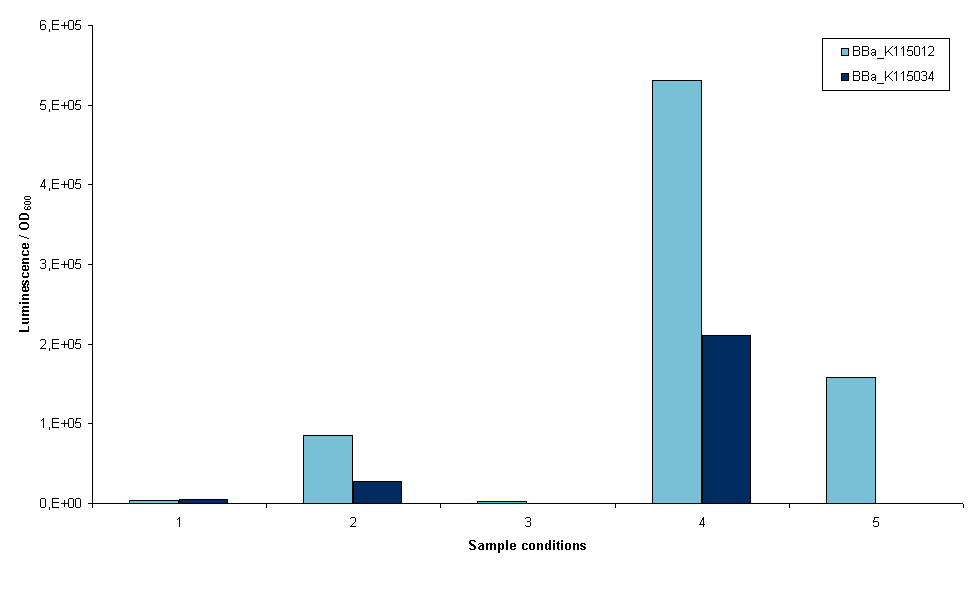
- 1. Induced growth overnight at room temperature, lysis by sonication
- 2. Induced growth overnight at 37ºC, lysis by sonication
- 3. Induced growth two times overnight at room temperature, lysis by sonication
- 4. Induced growth two times overnight at room temperature, lysis by Promega lysis buffer
- 5. Non-induced growth overnight at 37ºC, lysis by sonication (no induction at all, only BBa_K115012)
Several conclusions can be drawn from figure 3, although it has to be noted that this result is only one measurement. Furthermore, room temperature was not controlled. It may have fluctuated between 20 and 27ºC. The first conclusion is that we are able to measure luciferase with these constructs. This means that all parts used work correctly, including our ribosome binding site. The second conclusion is that a differential amount of luminescence is measured between strains BBa_K115012 and BBa_K115034, although in these results we cannot see a difference that is temperature dependent. This can be seen when the bars in conditions 1 and 2 are compared, BBa_K115012 gives a larger difference in luminescence from room temperature to 37ºC than BBa_K115034. The second conclusion is that luminescence measured when lysis is performed with the provided buffer is much higher (compare bars of condition 4 to the rest). The condition measured is not optimal as room temperature is used rather than 37ºC, but even now luminescence is a multitude higher than any of the sonicated samples. The reason for this is not clear: it could be that the lysis buffer lyses cells more efficiently, alternatively it could be that sonication denaturates a large part of the luciferase in the cell. Another conclusion is that the arabinose promoter does not work; a similar amount of luminescence was measured in samples where arabinose was added as compared to non-induced samples (figure 3, compare 5 to 1, 2, and 3).
Total protein content measurements in stead of OD measurements
To compare the amount of luciferase expression measured, the total luciferase expression should be normalized. In the first experiment we normalized the expression to the OD600 in the total culture. The OD600 is an indication for the amount of cells present in the sample. However in this set up, luciferase does not leave the cell. This means cell lysis should be performed under conditions that leave the luciferase protein intact for measurements. This poses a challenge: because the efficiency of lysis is unknown and cannot be presumed constant, we will need to perform additional protein content measurements.
 "
"