TUDelft/29 September 2008
From 2008.igem.org
| July | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| 21 | 22 | 23 | 24 | 25 | 26 | 27 |
| 28 | 29 | 30 | 31 | |||
| August | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | ||||
| 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| 18 | 19 | 20 | 21 | 22 | 23 | 24 |
| 25 | 26 | 27 | 28 | 29 | 30 | 31 |
| September | ||||||
| M | T | W | T | F | S | S |
| [http://2008.igem.org/TUDelft/1_September_2008 1] | [http://2008.igem.org/TUDelft/2_September_2008 2] | [http://2008.igem.org/TUDelft/3_September_2008 3] | [http://2008.igem.org/TUDelft/4_September_2008 4] | [http://2008.igem.org/TUDelft/5_September_2008 5] | [http://2008.igem.org/wiki/index.php?title=TUDelft/6_September_2008&action=edit 6] | [http://2008.igem.org/wiki/index.php?title=TUDelft/7_September_2008&action=edit 7] |
| [http://2008.igem.org/TUDelft/8_September_2008 8] | [http://2008.igem.org/TUDelft/9_September_2008 9] | [http://2008.igem.org/TUDelft/10_September_2008 10] | [http://2008.igem.org/TUDelft/11_September_2008 11] | [http://2008.igem.org/TUDelft/12_September_2008 12] | [http://2008.igem.org/wiki/index.php?title=TUDelft/13_September_2008&action=edit 13] | [http://2008.igem.org/wiki/index.php?title=TUDelft/14_September_2008&action=edit 14] |
| [http://2008.igem.org/TUDelft/15_September_2008 15] | [http://2008.igem.org/TUDelft/16_September_2008 16] | [http://2008.igem.org/TUDelft/17_September_2008 17] | [http://2008.igem.org/TUDelft/18_September_2008 18] | [http://2008.igem.org/TUDelft/19_September_2008 19] | [http://2008.igem.org/wiki/index.php?title=TUDelft/20_September_2008&action=edit 20] | [http://2008.igem.org/wiki/index.php?title=TUDelft/21_September_2008&action=edit 21] |
| [http://2008.igem.org/TUDelft/22_September_2008 22] | [http://2008.igem.org/TUDelft/23_September_2008 23] | [http://2008.igem.org/TUDelft/24_September_2008 24] | [http://2008.igem.org/TUDelft/25_September_2008 25] | [http://2008.igem.org/wiki/index.php?title=TUDelft/26_September_2008&action=edit 26] | [http://2008.igem.org/wiki/index.php?title=TUDelft/27_September_2008&action=edit 27] | [http://2008.igem.org/wiki/index.php?title=TUDelft/28_September_2008&action=edit 28] |
| [http://2008.igem.org/TUDelft/29_September_2008 29] | [http://2008.igem.org/TUDelft/30_September_2008 30] | |||||
| October | ||||||
| M | T | W | T | F | S | S |
| [http://2008.igem.org/TUDelft/1_October_2008 1] | [http://2008.igem.org/TUDelft/2_October_2008 2] | [http://2008.igem.org/TUDelft/3_October_2008 3] | [http://2008.igem.org/wiki/index.php?title=TUDelft/4_October_2008&action=edit 4] | [http://2008.igem.org/wiki/index.php?title=TUDelft/5_October_2008&action=edit 5] | ||
| [http://2008.igem.org/TUDelft/6_October_2008 6] | [http://2008.igem.org/TUDelft/7_October_2008 7] | [http://2008.igem.org/TUDelft/8_October_2008 8] | [http://2008.igem.org/TUDelft/9_October_2008 9] | [http://2008.igem.org/TUDelft/10_October_2008 10] | [http://2008.igem.org/wiki/index.php?title=TUDelft/11_October_2008&action=edit 11] | [http://2008.igem.org/wiki/index.php?title=TUDelft/12_October_2008&action=edit 12] |
| [http://2008.igem.org/TUDelft/13_October_2008 13] | [http://2008.igem.org/TUDelft/14_October_2008 14] | [http://2008.igem.org/TUDelft/15_October_2008 15] | [http://2008.igem.org/TUDelft/16_October_2008 16] | [http://2008.igem.org/TUDelft/17_October_2008 17] | [http://2008.igem.org/wiki/index.php?title=TUDelft/18_October_2008&action=edit 18] | [http://2008.igem.org/wiki/index.php?title=TUDelft/19_October_2008&action=edit 19] |
| [http://2008.igem.org/TUDelft/20_October_2008 20] | [http://2008.igem.org/TUDelft/21_October_2008 21] | [http://2008.igem.org/TUDelft/22_October_2008 22] | [http://2008.igem.org/TUDelft/23_October_2008 23] | [http://2008.igem.org/TUDelft/24_October_2008 24] | [http://2008.igem.org/wiki/index.php?title=TUDelft/25_October_2008&action=edit 25] | [http://2008.igem.org/wiki/index.php?title=TUDelft/26_October_2008&action=edit 26] |
| [http://2008.igem.org/wiki/index.php?title=TUDelft/27_October_2008&action=edit 27] | [http://2008.igem.org/wiki/index.php?title=TUDelft/28_October_2008&action=edit 28] | [http://2008.igem.org/wiki/index.php?title=TUDelft/29_October_2008&action=edit 29] | [http://2008.igem.org/wiki/index.php?title=TUDelft/30_October_2008&action=edit 30] | [http://2008.igem.org/wiki/index.php?title=TUDelft/31_October_2008&action=edit 31] | ||
September 29th
Touchdown PCR
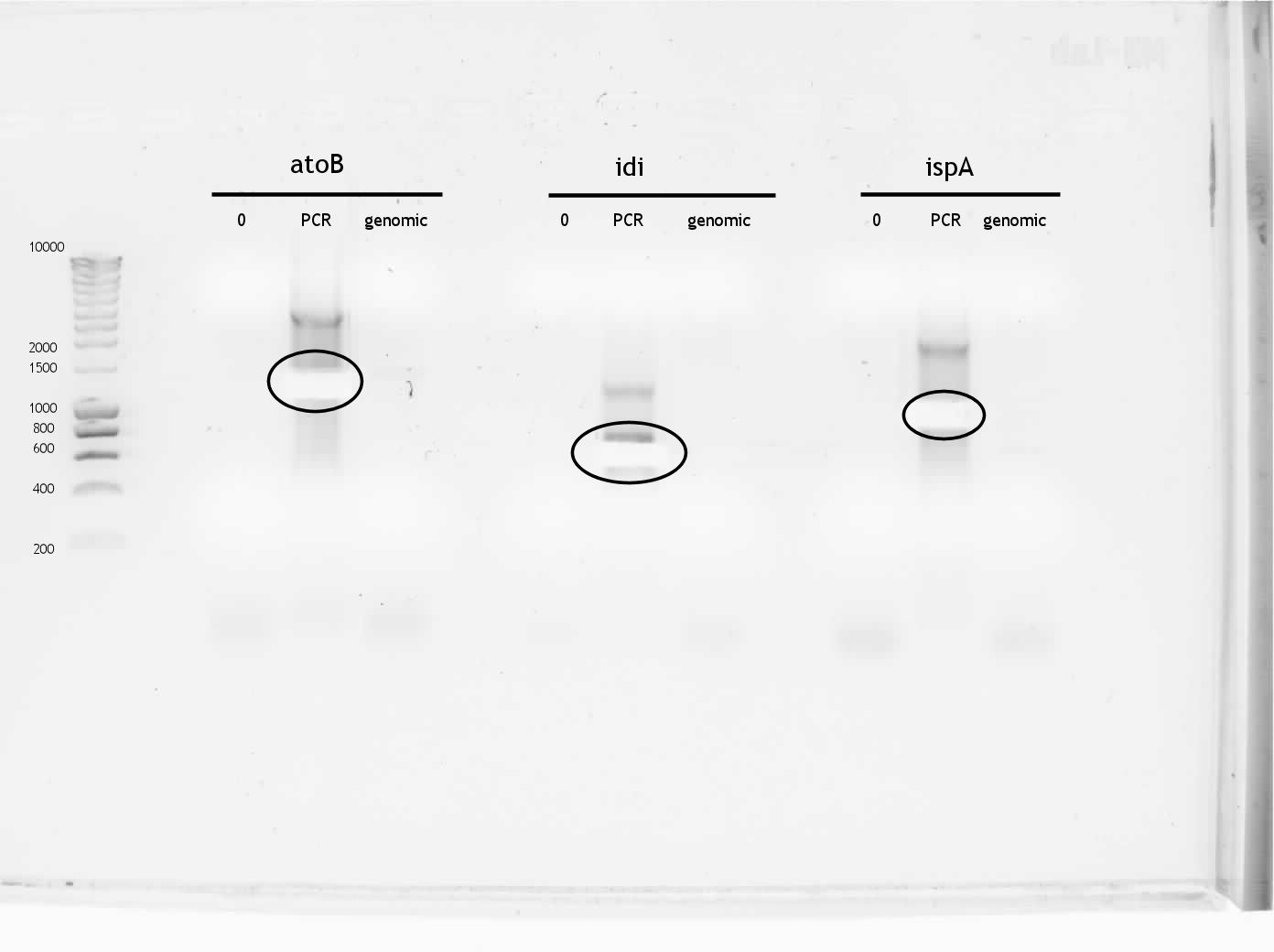
Because the new primers have long sequences that are not supposed to bind to the target DNA (the pre- and suffix for forward and reverse primer, respectively), a low annealing temperature could lead to a lot of aspecific product formation. To suppress this, a touchdown PCR was performed with the new primers on the PCR products of atoB, idi and ispA and genomic DNA of E. coli. In both cases 60 ng of template DNA and the platinum Pfx enzyme was used. The PCR program looked like this:
1. 5' @ 94°C
2. 1' @ 94°C
3. 1' @ 65°C --> temperature is lowered with 0.5°C per cycle
4. 3' @ 72°C
5. go to 2, 20 cycles in total
6. 1' @ 94°C
7. 1' @ 94°C
8. 3' @ 72°C
9. go to 6, 20 cycles in total
10. 7' @ 72°C
11. ∞ @ 10°C
The products were put on gel:
Circled areas indicate cut bands, after the picture was taken some extra agarose in the idi lane was removed. Expected sizes were for atoB ~1200bp, idi ~580bp and ispA ~975bp
As can be seen, the PCR did not succeed for the genomic DNA, but it did work on the PCR products. The reason for this is probably that although there is an even amount of template DNA in both cases, there is of course a lot more specific DNA binding spots in the PCR product. The gel bands were cut (see figure) and DNA isolated using a Qiagen kit. The 260/280 was around 1.7 and DNA content around 21 ng/ul for all products (atoB, idi and ispA) according to NanoDrop.
Growing for midiprep
K115022 and K115027 are incubated o/n @ 37 degrees Celcius.
 "
"