|
← Yesterday ↓ Calendar ↑Tomorrow →
Cloning of FlhB promoter
Protocol
- Preparation of the template :
Resuspension of 1 colony E.coli K12 strain MG 1655 in 100µl of water.
1µl of dNTP
10µl Buffer Phusion 5X
2.5µl O 108
2.5µl O 109
1µl Template
1µl Phusion
33µL pure water
Result
| Well
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
|
| Sample
| 100 bp ladder
|
|
|
|
|
|
|
|
|
|
| Expected size (pb)
|
|
|
|
|
|
|
|
|
| Measured size (pb)
|
|
|
|
|
|
|
|
|
|
|
Construction for FIFO
Aim : Construction of pFlgA - YFP tripart (+/- LVA) "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432)     
Digestion
Digestion
Protocol Digestion
| Name
| Template DNA
| Description
| Vol MP (µl)
| Vol H2O (µl)
| Enzymes
|
| D166
| MP165.1
| RBS+ YFP LVA- term - FV
| 7.63
| 17
| EcoRI and XbaI
|
| D167
| MP166.1
| RBS+ YFP LVA+ term - FV
| 8.9
| 15.8
| EcoRI and XbaI
|
Protocol
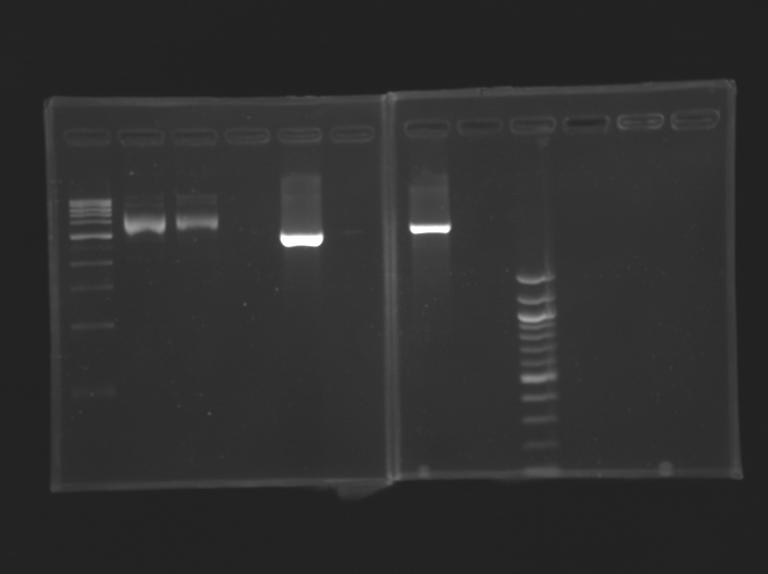 Gel Extraction of D166-D167
| Well
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
|
| Sample
| 1kb
ladder
| MP165.1
| MP166.1
| no sample
| D166
| no sample
| D167
| no sample
| 100pb
ladder
| no sample
|
| Expected size (pb)
|
| 2 957
| 2 996
|
| 2942
|
| 2981
|
|
| Measured size (pb)
|
| 3 000
| 3 000
|
| 2 800
|
| 3 000
|
|
Measurement of the concentration of D166 & D167 purified
Protocol (it's same that for Miniprep)
| Digestion Name
| Concentration (µg/mL)
| Ratio 260/280
|
| D166
| 11
| 4.73
|
| D167
| 10
| 2.84
|
Ligation
Protocol
| Ligation Name
| Vector Name
| Volume Vector (µL)
| Insert
| Volume Insert (µL)
|
| L160
| D166
| 3.63
| D132
| 2.03
|
| L161
| D167
| 4.00
| D132
| 2.01
|
| Control L160
| D166
| 3.63
| -
| -
|
| Control L161
| D167
| 4.00
| -
| -
|
Screening of the cloning of E0240 and FlhDC+promotor
We obtained colonies isolated with the dilution 1/100; 13 clones were analysed by PCR
PCR screening
reaction mixture (25 µL)
- 12,5 µL Quick load PCR Mixture 2X
- 0,5 µL O18
- 0,5 µL O19
- 11,5 µL water
PCR screening programm
- elongation time: 1 min 30
- primers: O18 and O19
- positive control: S158 (pSB3K3)
- negative control: no template
Electrophoresis
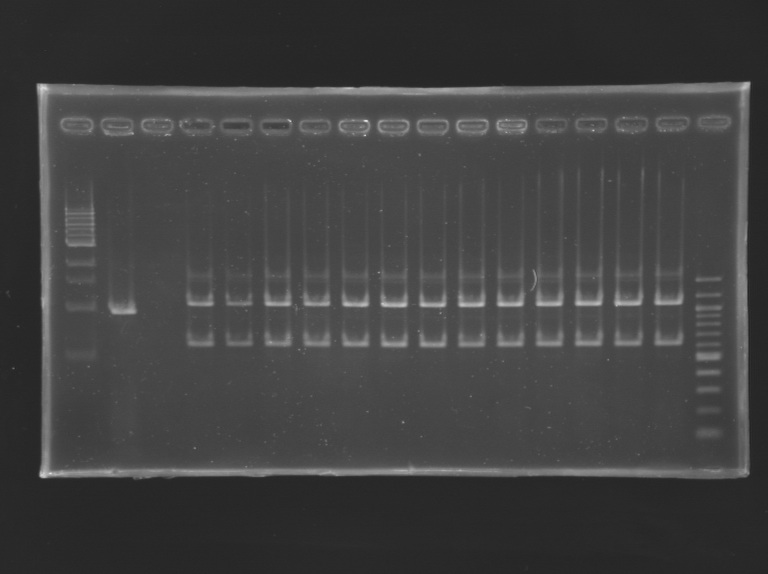 Gel n°1 (E0240 screening)  Gel n°2 (FlhDC+promotor screening) - 1% agarose gel
- 10 µL loaded
| Gel n°1 (E0240 in pSB3K3)
|
| well n°
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
| 13
| 14
| 15
| 16
| 17
|
| sample
| 1 kb
DNA ladder
| control +
| control -
| S159.1
| 100 bp
DNA ladder
|
| colonie n°
|
|
|
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
| 13
|
|
| expected size
|
|
|
| 1192 bp
|
|
| measured size
|
|
|
| 1,5 kb
1,1 kb
0,6 kb
|
|
| Gel n°2 (FlhDC+promotor in pSB1A2)
|
| well n°
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
| 13
| 14
| 15
| 16
| 17
|
| sample
| 1 kb
DNA ladder
| control +
| control -
| S161.1
| 100 bp
DNA ladder
|
| colonie n°
|
|
|
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
| 13
|
|
| expected size
|
|
|
| 1403 bp
|
|
| measured size
|
|
|
| 0,3 kb
|
|
Results:
*The clone of E0240 (S159.1) always have several bands amplified by PCR.
It might contain different plasmids.
*The clone of FlhDC+promotor (S161.1) don't have the correct size band.
It also doesn't have the insert in the plasmid.
Construction for synchronization
Ligations
Protocol
| Ligation Name
| Description
| Vector Name
| Volume vector (µL)
| Insert Name
| Volume insert(µL)
|
| L158
| rbs-TetR + gfp tripart
     
| D110 (BV)
| 2
| D131 (BI)
| 2.89
|
| control L158
| rbs-TetR + gfp tripart
     
| D110 (BV)
| 2
|
|
|
| L159
| rbs-lasI
 
| D125 (FV)
| 2.08
| D109 (FI)
| 1.15
|
| control L159
| rbs-lasI
 
| D125 (FV)
| 2.08
|
|
|
Promoter characterization plasmids
Ligations from digestions from 20th
Top 10 cells were used
Protocol
| Ligation name
| Vector digestion
| Vector description
| Vector conc. ug/mL
| Vector volume
| Insert digestion
| Insert description
| Insert conc. ug/mL
| Insert volume
| Product description
| Antibiotic
|
| L155
| D164
| J23101 promoter
| 3
| 16
| D163
| gfp generator
| 14
| 8
| J23101 promoter-gfp generator
| Amp
|
| L156
| D161
| pTet promoter
| 39
| 1.25
| D163
| gfp generator
| 14
| 5
| pTet promoter-gfp generator
| Kana
|
| Control L156
| D161
|
|
| 1.25
| none
|
|
|
| Vector autoligation control
| Kana
|
| L157
| D125.2
| B0015
| 15
| 3
| D162
| tetR
| 11
| 4
| tetR-B0015
| Kana
|
| Control L157
| D125.2
|
|
| 3
| none
|
|
|
| Vector autoligation control
| Kana
|
| L166
| D185
| RBS B0032
| 21
| 2
| D182
| tetR
| 10
| 6.5
| RBS B0032 - tetR
| Amp
|
| Control L166
| D185
|
|
| 2
| none
|
|
|
| Vector autoligation control
| Amp
|
| L167
| D181
| pTet
| 1
| 14
| D184
| gfp generator
| 14
| 3
| gfp generator - pTet
| Amp
|
| Control L167
| D181
| pTet
|
| 14
| none
|
|
|
| Vector autoligation control
| Amp
|
|
 "
"



