Team:Paris/August 11
From 2008.igem.org
(→PCR verification/Analysis) |
(→Culture of ligation transformants) |
||
| (8 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
==Transformation== | ==Transformation== | ||
| + | All the ligations were transformed according [[Team:Paris/Notebook/Protocols#Transformation|transformation for Top10 protocol]] | ||
| + | |||
| - | |||
| - | |||
==PCR== | ==PCR== | ||
| Line 120: | Line 120: | ||
|MP143 | |MP143 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_130' |
| - | | - | ||
|O141_O140 | |O141_O140 | ||
|Water | |Water | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_131 |
|flhD RBS- | |flhD RBS- | ||
|O132_O133 | |O132_O133 | ||
|MG1655 | |MG1655 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_131' |
| - | | - | ||
|O132_O133 | |O132_O133 | ||
|Water | |Water | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_132 |
|flhC RBS- | |flhC RBS- | ||
|O130_O131 | |O130_O131 | ||
|MG1655 | |MG1655 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_132' |
| - | | - | ||
|O130_O131 | |O130_O131 | ||
|Water | |Water | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_133 |
|flhDC + prom | |flhDC + prom | ||
|O110_O131 | |O110_O131 | ||
|MG1655 | |MG1655 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_133' |
| - | | - | ||
|O110_O131 | |O110_O131 | ||
|Water | |Water | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_134 |
|flhDC + prom | |flhDC + prom | ||
|O111_O131 | |O111_O131 | ||
|MG1655 | |MG1655 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_134' |
| - | | - | ||
|O111_O131 | |O111_O131 | ||
|Water | |Water | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_135 |
|pfliL | |pfliL | ||
|O124_O125 | |O124_O125 | ||
|MG1655 | |MG1655 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_135' |
| - | | - | ||
|O124_O125 | |O124_O125 | ||
|Water | |Water | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_136 |
|pflhDC | |pflhDC | ||
|O110_O113 | |O110_O113 | ||
|MG1655 | |MG1655 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_136' |
| - | | - | ||
|O110_O113 | |O110_O113 | ||
|Water | |Water | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_137 |
|pflhDC | |pflhDC | ||
|O111_O113 | |O111_O113 | ||
|MG1655 | |MG1655 | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_137' |
| - | | - | ||
|O111_O113 | |O111_O113 | ||
| Line 230: | Line 230: | ||
|style="background: #cbff7B"|<center> 900 bp </center> | |style="background: #cbff7B"|<center> 900 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_130' |
|Negative Control | |Negative Control | ||
|Gel 1 | |Gel 1 | ||
| Line 237: | Line 237: | ||
|style="background: #cbff7B"|<center> 0 bp </center> | |style="background: #cbff7B"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_131 |
|flhD RBS- | |flhD RBS- | ||
|Gel 1 | |Gel 1 | ||
| Line 244: | Line 244: | ||
|style="background: #cbff7B"|<center> 350 bp </center> | |style="background: #cbff7B"|<center> 350 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_131' |
|Negative Control | |Negative Control | ||
|Gel 1 | |Gel 1 | ||
| Line 251: | Line 251: | ||
|style="background: #cbff7B"|<center> 0 bp </center> | |style="background: #cbff7B"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_132 |
|flhC RBS- | |flhC RBS- | ||
|Gel 1 | |Gel 1 | ||
| Line 258: | Line 258: | ||
|style="background: #cbff7B"|<center> 600 bp </center> | |style="background: #cbff7B"|<center> 600 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_132' |
|Negative Control | |Negative Control | ||
|Gel 1 | |Gel 1 | ||
| Line 265: | Line 265: | ||
|style="background: #cbff7B"|<center> 0 bp </center> | |style="background: #cbff7B"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_133 |
|flhDC + prom | |flhDC + prom | ||
|Gel 2 | |Gel 2 | ||
| Line 272: | Line 272: | ||
|style="background: #cbff7B"|<center> 1300 bp </center> | |style="background: #cbff7B"|<center> 1300 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_133' |
|Negative Control | |Negative Control | ||
|Gel 2 | |Gel 2 | ||
| Line 279: | Line 279: | ||
|style="background: #cbff7B"|<center> 0 bp </center> | |style="background: #cbff7B"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_134 |
|flhDC + prom | |flhDC + prom | ||
|Gel 2 | |Gel 2 | ||
| Line 286: | Line 286: | ||
|style="background: #ff6d73"|<center> 2100 pb </center> | |style="background: #ff6d73"|<center> 2100 pb </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_134' |
|Negative Control | |Negative Control | ||
|Gel 2 | |Gel 2 | ||
| Line 295: | Line 295: | ||
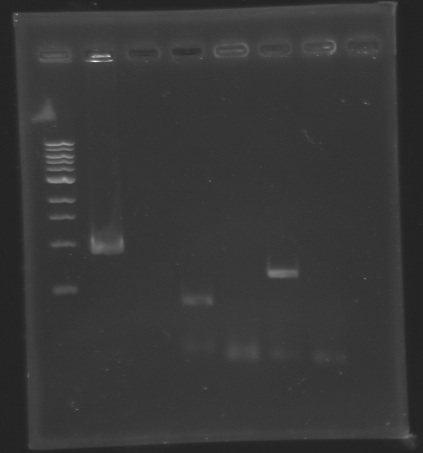
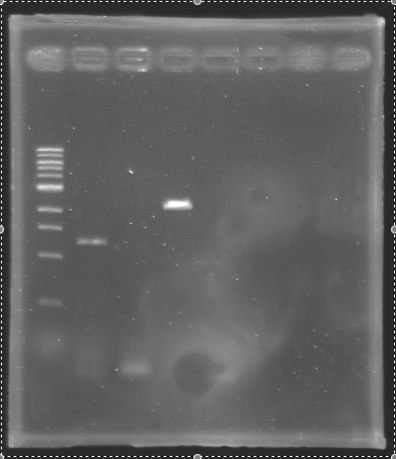
| - | ==> '''Conclusion :''' We observed the size expected for the PCR products, but not for pflhDC ( | + | ==> '''Conclusion :''' We observed the size expected for the PCR products, but not for pflhDC (PCR_134), is right. We hypothesis for PCR_138 that the size is longer that expected due to the aspecific fixation of Oligo O111 (upper to the real site). |
| Line 315: | Line 315: | ||
|align="center"|'''Measured size''' | |align="center"|'''Measured size''' | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_135 |
|pfliL | |pfliL | ||
|Gel 3 | |Gel 3 | ||
| Line 322: | Line 322: | ||
|style="background: #ff6d73"|<center> 0 bp </center> | |style="background: #ff6d73"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_135' |
|Negative Control | |Negative Control | ||
|Gel 3 | |Gel 3 | ||
| Line 329: | Line 329: | ||
|style="background: #cbff7B"|<center> 0 bp </center> | |style="background: #cbff7B"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_136 |
|pflhDC | |pflhDC | ||
|Gel 3 | |Gel 3 | ||
| Line 336: | Line 336: | ||
|style="background: #ff6d73"|<center> 0 bp </center> | |style="background: #ff6d73"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_136' |
|Negative Control | |Negative Control | ||
|Gel 3 | |Gel 3 | ||
| Line 343: | Line 343: | ||
|style="background: #cbff7B"|<center> 0 bp </center> | |style="background: #cbff7B"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_137 |
|pflhDC | |pflhDC | ||
|Gel 3 | |Gel 3 | ||
| Line 350: | Line 350: | ||
|style="background: #ff6d73"|<center> 0 bp </center> | |style="background: #ff6d73"|<center> 0 bp </center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |PCR_137' |
|Negative Control | |Negative Control | ||
|Gel 3 | |Gel 3 | ||
| Line 359: | Line 359: | ||
| - | ==> '''Conclusion :''' We need to | + | ==> '''Conclusion :''' We need to repeat the experiments. |
| - | ==Culture of ligation transformants== | + | ==Culture of ligation transformants (pFlgA, pFlgB and pFlhB)== |
*4 clones of each transformation were cultured in '''7,5 mL LB + ampicilline'''. The colonies picked up were the rest of those already picked up from the transformation plate (for the PCR screening of August 8). | *4 clones of each transformation were cultured in '''7,5 mL LB + ampicilline'''. The colonies picked up were the rest of those already picked up from the transformation plate (for the PCR screening of August 8). | ||
Latest revision as of 16:11, 18 August 2008
TransformationAll the ligations were transformed according transformation for Top10 protocol
PCRWe performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR amplificationProtocol
For each sample, 1 µl dNTP
PCR verification/AnalysisAfter the PCR :
ladder : 10µl ladder 1 kb
ladder : 10µl ladder 100 bp
Culture of ligation transformants (pFlgA, pFlgB and pFlhB)
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"