Team:Paris/August 20
From 2008.igem.org
(Difference between revisions)
(→Results of the transformation we did yesterday) |
(→Screening) |
||
| (71 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Calendar_Links|August 19|August 21}} | {{Paris/Calendar_Links|August 19|August 21}} | ||
| - | =Screening of the cloning of E0240 and FlhDC+promotor= | + | ='''Screening of the cloning of E0240 and FlhDC+promotor'''= |
==Spreading the clones in order to obtain single colonies== | ==Spreading the clones in order to obtain single colonies== | ||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| Line 35: | Line 35: | ||
*Overnight incubation (37°C) | *Overnight incubation (37°C) | ||
| + | ='''Miniprep and stock glycerol of stable strains with biobricks 2008'''= | ||
| - | |||
| - | |||
| - | |||
*[[Team:Paris/Notebook/Protocols#Glycerol stocks| Protocol stocks]] | *[[Team:Paris/Notebook/Protocols#Glycerol stocks| Protocol stocks]] | ||
| - | + | *[[Team:Paris/Notebook/Protocols#Minipreps (Kit_Qiagen)| Protocol Miniprep]] | |
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
|'''Stock number''' | |'''Stock number''' | ||
| + | |'''Miniprep number''' | ||
|'''Biobricks''' | |'''Biobricks''' | ||
|'''Description''' | |'''Description''' | ||
|- | |- | ||
|S163.1 | |S163.1 | ||
| + | |MP163.1 | ||
|rowspan="2"| B0032 | |rowspan="2"| B0032 | ||
| - | |rowspan="2"| RBS | + | |rowspan="2"| [[Image:Icon_rbs.png]]<br>RBS |
|- | |- | ||
|S163.2 | |S163.2 | ||
| + | |MP163.2 | ||
|- | |- | ||
|S164.1 | |S164.1 | ||
| + | |MP164.1 | ||
|rowspan="2"| E0422 | |rowspan="2"| E0422 | ||
| - | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]RBS+ ECFP+ LVA+ term | + | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>RBS+ ECFP+ LVA+ term |
|- | |- | ||
|S164.2 | |S164.2 | ||
| + | |MP164.2 | ||
|- | |- | ||
|S165.1 | |S165.1 | ||
| + | |MP165.1 | ||
|rowspan="2"| E0430 | |rowspan="2"| E0430 | ||
| - | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]RBS+ YFP+ LVA- term | + | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>RBS+ YFP+ LVA- term |
|- | |- | ||
|S165.2 | |S165.2 | ||
| + | |MP165.2 | ||
|- | |- | ||
|S166.1 | |S166.1 | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|MP166.1 | |MP166.1 | ||
|rowspan="2"| E0432 | |rowspan="2"| E0432 | ||
| - | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]RBS+ YFP LVA+ term | + | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>RBS+ YFP LVA+ term |
|- | |- | ||
| + | |S166.2 | ||
|MP166.2 | |MP166.2 | ||
|- | |- | ||
| + | |S167.1 | ||
|MP167.1 | |MP167.1 | ||
|rowspan="2"| E0420 | |rowspan="2"| E0420 | ||
| - | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]RBS+ ECFP LVA- term | + | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>RBS+ ECFP LVA- term |
|- | |- | ||
| + | |S167.2 | ||
|MP167.2 | |MP167.2 | ||
|- | |- | ||
| + | |S168.1 | ||
|MP168.1 | |MP168.1 | ||
|rowspan="2"| I732078 | |rowspan="2"| I732078 | ||
| - | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]RBS+ mRFP LVA+ term | + | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>RBS+ mRFP LVA+ term |
|- | |- | ||
| + | |S168.2 | ||
|MP168.2 | |MP168.2 | ||
|} | |} | ||
| - | + | ='''Construction for FIFO'''= | |
| - | + | ||
| - | + | Aim : Construction of pFlgA - YFP tripart (+/- LVA) ''' "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432)''' [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Digestion== | ==Digestion== | ||
| Line 353: | Line 168: | ||
|1.63 | |1.63 | ||
|} | |} | ||
| - | |||
===Digestion=== | ===Digestion=== | ||
| Line 368: | Line 182: | ||
|D166 | |D166 | ||
|MP165.1 | |MP165.1 | ||
| - | | - FV | + | | RBS+ YFP LVA- term - FV |
|7.63 | |7.63 | ||
|17 | |17 | ||
| Line 375: | Line 189: | ||
|D167 | |D167 | ||
|MP166.1 | |MP166.1 | ||
| - | | - | + | | RBS+ YFP LVA+ term - FV |
|8.9 | |8.9 | ||
|15.8 | |15.8 | ||
| Line 382: | Line 196: | ||
|D131 | |D131 | ||
|MP122.2 | |MP122.2 | ||
| - | | - | + | | GFP tripart - I |
|10.2 | |10.2 | ||
|14.5 | |14.5 | ||
|XbaI and PstI | |XbaI and PstI | ||
|} | |} | ||
| - | |||
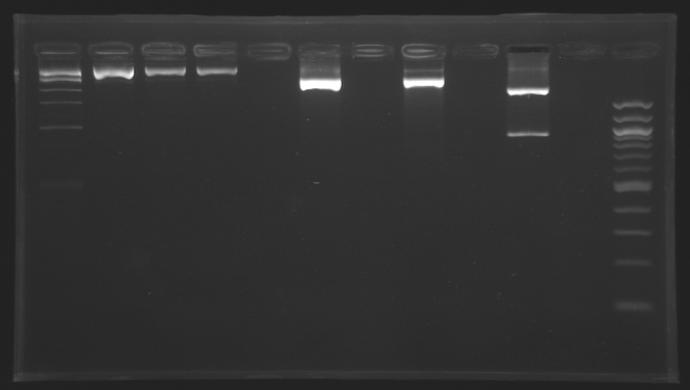
==='''Gel Extraction'''=== | ==='''Gel Extraction'''=== | ||
| - | |||
[[Team:Paris/Notebook/Protocols#Migration after extraction |Protocol]] | [[Team:Paris/Notebook/Protocols#Migration after extraction |Protocol]] | ||
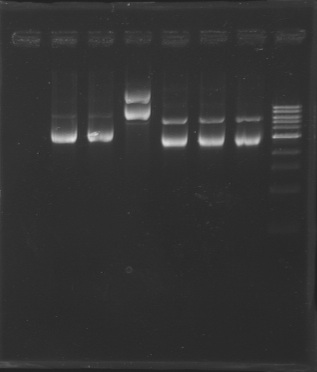
| - | + | [[Image:KR000193b.jpg| thumb|Gel Extraction of D166-D167-D131]] | |
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
|'''Well''' | |'''Well''' | ||
| - | |||
| - | |||
| - | |||
| - | |||
|1 | |1 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|2 | |2 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|3 | |3 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|4 | |4 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|5 | |5 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|6 | |6 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|7 | |7 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|8 | |8 | ||
| - | | | + | |9 |
| - | | | + | |10 |
| - | | | + | |11 |
| + | |12 | ||
|- | |- | ||
| - | | | + | |'''Sample''' |
| + | |1kb ladder | ||
| + | |MP165.1 | ||
| + | |MP166.1 | ||
| + | |MP122.2 | ||
| + | |no sample | ||
| + | |D166 | ||
| + | |no sample | ||
| + | |D167 | ||
|no sample | |no sample | ||
| - | |||
| - | |||
| - | |||
| - | |||
|D131 | |D131 | ||
| - | | | + | |no sample |
| - | | | + | |100pb ladder |
|- | |- | ||
| - | | | + | |'''Expected size (pb)''' |
| - | + | ||
| | | | ||
| + | |2 957 | ||
| + | |2 996 | ||
| + | |2 957 | ||
| | | | ||
| + | |2942 | ||
| + | | | ||
| + | |914 | ||
| + | | | ||
| + | |900 | ||
|- | |- | ||
| - | | | + | |'''Measured size (pb)''' |
| - | | | + | | |
| + | |style="background: #cbff7B"| 3 000 | ||
| + | |style="background: #cbff7B"| 3 000 | ||
| + | |style="background: #cbff7B"| 3 000 | ||
| + | | | ||
| + | |style="background: #ff6d73"| 2500 | ||
| + | | | ||
| + | |style="background: #ff6d73"| 2500 | ||
| + | | | ||
| + | |style="background: #cbff7B"| 950 | ||
| | | | ||
| | | | ||
|} | |} | ||
| + | => Following a mistake, the right E0430(D166) and E0432(D167) digestion, have not been purified. <br>So we need to repeat the same digestion experiment tomorrow morning. | ||
| - | |||
| - | |||
| - | |||
| + | ='''Construction of: promotor-rbs-LasR-dbl ter'''= | ||
| + | ==Results of the transformation we did [[Team:Paris/August 18 |the day before yesterday]]== | ||
| + | ===Number of colonies=== | ||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
|'''Ligation name''' | |'''Ligation name''' | ||
| Line 514: | Line 316: | ||
|} | |} | ||
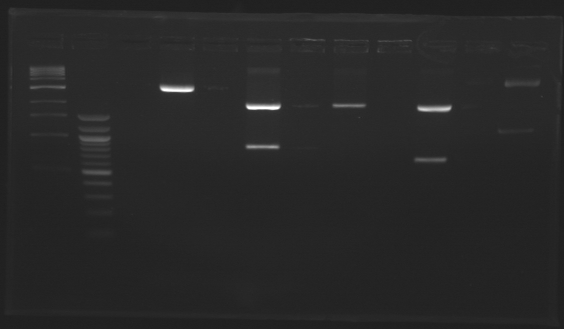
| - | + | ==PCR Screening== | |
| - | + | [[Team:Paris/Notebook/Protocols#PCR Screening| Protocol]] | |
| - | + | [[Image:J23100-J23101+D165.jpg |thumb| Ligations results J23100+D165 and J23101+D165]] | |
| - | [[Image: | + | |
| - | + | ||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
|'''Well''' | |'''Well''' | ||
| - | |||
| - | |||
| - | |||
| - | |||
|1 | |1 | ||
| - | |||
| - | |||
| - | |||
| - | |||
|2 | |2 | ||
| - | |||
| - | |||
|3 | |3 | ||
| - | |||
| - | |||
|4 | |4 | ||
| - | |||
| - | |||
|5 | |5 | ||
| - | |||
| - | |||
|6 | |6 | ||
| - | |||
| - | |||
|7 | |7 | ||
| - | |||
| - | |||
|8 | |8 | ||
| - | |||
| - | |||
|9 | |9 | ||
| + | |10 | ||
| + | |11 | ||
| + | |12 | ||
| + | |13 | ||
| + | |14 | ||
| + | |15 | ||
| + | |16 | ||
| + | |17 | ||
| + | |- | ||
| + | |'''Sample''' | ||
| + | |colspan="8" |L153 | ||
|1kb ladder | |1kb ladder | ||
| + | |colspan="8" |L154 | ||
| + | |- | ||
| + | |'''Clone''' | ||
| + | |.1 | ||
| + | |.2 | ||
| + | |.3 | ||
| + | |.4 | ||
| + | |.5 | ||
| + | |.6 | ||
| + | |.7 | ||
| + | |.8 | ||
| | | | ||
| + | |.1 | ||
| + | |.2 | ||
| + | |.3 | ||
| + | |.4 | ||
| + | |.5 | ||
| + | |.6 | ||
| + | |.7 | ||
| + | |.8 | ||
| + | |- | ||
| + | |'''Expected size''' | ||
| + | |colspan="8" |1194 | ||
| | | | ||
| + | |colspan="8" |1194 | ||
|- | |- | ||
| - | | | + | |'''Measured size''' |
| + | |style="background: #cbff7B" colspan="8" |1200 | ||
| + | | | ||
| + | |style="background: #cbff7B" colspan="8" | 1200 | ||
| + | |} | ||
| + | |||
| + | ==Minipreps== | ||
| + | {|border="1" style="text-align: center" | ||
| + | |'''Miniprep Name''' | ||
| + | |'''Ligation name''' | ||
| + | |'''Antibio''' | ||
| + | |'''Biobricks''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |MP169.1 | ||
| + | |L153.1 | ||
| + | |rowspan="4" |Amp | ||
| + | |rowspan="2" |[[Image:part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_coding.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| + | |rowspan="2" |J23100 - rbs-LasR-Double terminator | ||
| + | |- | ||
| + | |MP169.2 | ||
| + | |L153.2 | ||
| + | |- | ||
| + | |MP170.1 | ||
|L154.1 | |L154.1 | ||
| - | |rowspan=" | + | |rowspan="2" |[[Image:part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_coding.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
| - | |rowspan=" | + | |rowspan="2" |J23101 - rbs-LasR - Double terminator |
|- | |- | ||
| - | | | + | |MP170.2 |
|L154.2 | |L154.2 | ||
| + | |} | ||
| + | |||
| + | |||
| + | ='''Promoter characterization plasmids'''= | ||
| + | ==Ligation== | ||
| + | Our ligations from yesterday didn't work. The positive control for transformation worked. | ||
| + | |||
| + | ==Digestion== | ||
| + | We had a problem with a gel extraction so we have to make again the digestions from yesterday, Other digestions made: | ||
| + | |||
| + | [[Team:Paris/Notebook/Protocols#Digestion|Protocol Digestion]] | ||
| + | |||
| + | {| border="1" style="text-align: center" | ||
| + | |'''Digestion name''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |'''Miniprep used''' | ||
| + | |'''Enzymes''' | ||
| + | |'''Concentration after gel extraction''' | ||
|- | |- | ||
| - | | | + | |D179 |
| - | | | + | |MP3.4 |
| + | |B0015 (double terminator B0010-B0012) - BV | ||
| + | |4 | ||
| + | |SpeI and PstI | ||
| + | |9 | ||
|- | |- | ||
| + | |D180 | ||
| + | |MP101.1 | ||
| + | |promoter J23101- BV | ||
| + | |1 | ||
| + | |SpeI and PstI | ||
| + | |7 | ||
| + | |- | ||
| + | |D181 | ||
| + | |MP104.2 | ||
| + | |PTet (TetR repressible promoter) - FV | ||
| + | |1 | ||
| + | |EcoRI and XbaI | ||
| + | |1 | ||
| + | |- | ||
| + | |D182 | ||
| + | |MP114.1 | ||
| + | |TetR - BI | ||
| + | |1 | ||
| + | |XbaI and PstI | ||
| + | |10 | ||
| + | |- | ||
| + | |D183 | ||
| + | |MP119.3 | ||
| + | |pBad promoter - BI | ||
| + | |1 | ||
| + | |XbaI and PstI | ||
| + | |0 | ||
| + | |- | ||
| + | |D184 | ||
| + | |MP143.1 | ||
| + | |gfp generator - FI | ||
| + | |2 | ||
| + | |EcoRI and SpeI | ||
|13 | |13 | ||
| - | |||
|- | |- | ||
| - | | | + | |D185 |
| - | | | + | |MP163.1 |
| + | |B0032 RBS - BV | ||
| + | |2 | ||
| + | |SpeI and PstI | ||
| + | |21 | ||
| + | |} | ||
| + | |||
| + | |||
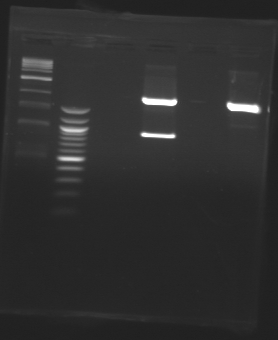
| + | D179 | ||
| + | D180 | ||
| + | D181 | ||
| + | D182 | ||
| + | D183 | ||
| + | [[Image:20-08-08.png|200px]] | ||
| + | D184 | ||
| + | D185 | ||
| + | [[Image:20-08-08bis.png|100px]] | ||
| + | |||
| + | |||
| + | ='''Sequencing'''= | ||
| + | *We received results of our sequencing from COCHIN | ||
| + | *We succeed for FlgA promoter | ||
| + | *FlgB and flhB don't match with our expected sequences.<br>We decided to cut our Miniprep product with other enzymes to check our sequence. | ||
| + | |||
| + | ==Digestion== | ||
| + | [[Team:Paris/Notebook/Protocols#Digestion |Protocol]] | ||
| + | *D176 : pFlgB digested with ApoI | ||
| + | *D177 : pFlgB digested with NruI | ||
| + | *D178 : pFlhB digested with BstAPI | ||
| + | |||
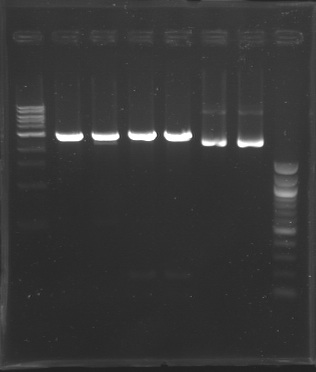
| + | ==Screening== | ||
| + | Gel 1 [[Image:Gel1 FlgB.jpg |150px]] Gel 2[[Image:Gel_2_FlhB.2.jpg |150px]] | ||
| + | |||
| + | |||
| + | {|border="1" style="text-align: center" | ||
| + | | | ||
| + | |colspan="8"|'''Gel 1''' | ||
| + | |colspan="9"|'''Gel 2''' | ||
|- | |- | ||
| - | | | + | |'''Well''' |
| - | | | + | |1 |
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |7 | ||
| + | |8 | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |7 | ||
| + | |8 | ||
|- | |- | ||
| - | | | + | |'''Sample''' |
| - | | | + | |1kb<br>ladder |
| + | |D176.1 | ||
| + | |D176.2 | ||
| + | |D176.3 | ||
| + | |D176.4 | ||
| + | |D177.1 | ||
| + | |D177.2 | ||
| + | |100pb<br>ladder | ||
| + | |no sample | ||
| + | |D177.3 | ||
| + | |D177.4 | ||
| + | |D178.1 | ||
| + | |D178.2 | ||
| + | |D178.3 | ||
| + | |D178.4 | ||
| + | |1kb<br>ladder | ||
|- | |- | ||
| - | | | + | |'''Expected size''' |
| - | | | + | | |
| + | |colspan="4" |2970<br>194<br>48 | ||
| + | |colspan="2" |3212 | ||
| + | | | ||
| + | |colspan="2" |3212 | ||
| + | |colspan="5" |3213 | ||
| + | |- | ||
| + | |'''Measured size''' | ||
| + | | | ||
| + | |style="background: #cbff7B" colspan="4" |2900<br>200<br>/ | ||
| + | |style="background: #ff6d73" colspan="2" |Plamsid not digested | ||
| + | | | ||
| + | |style="background: #ff6d73" colspan="7" |Plamsid not digested | ||
| + | | | ||
|} | |} | ||
| + | |||
| + | Conclusion => the sequence of pFlgB and pFlhB are not good we will tried to isolate pFlhB again. | ||
Latest revision as of 14:03, 6 September 2008
Screening of the cloning of E0240 and FlhDC+promotorSpreading the clones in order to obtain single colonies
The plates obtained from the speading of yesterday can't be used because there are not single colonies.
Miniprep and stock glycerol of stable strains with biobricks 2008Construction for FIFOAim : Construction of pFlgA - YFP tripart (+/- LVA) "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432) DigestionMeasurement of concentration of minipreps
Digestion
Gel Extraction
=> Following a mistake, the right E0430(D166) and E0432(D167) digestion, have not been purified.
Construction of: promotor-rbs-LasR-dbl terResults of the transformation we did the day before yesterdayNumber of colonies
PCR Screening
Minipreps
Promoter characterization plasmidsLigationOur ligations from yesterday didn't work. The positive control for transformation worked. DigestionWe had a problem with a gel extraction so we have to make again the digestions from yesterday, Other digestions made:
Sequencing
*We succeed for FlgA promoter *FlgB and flhB don't match with our expected sequences. Digestion
Screening
Conclusion => the sequence of pFlgB and pFlhB are not good we will tried to isolate pFlhB again. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"