Team:Paris/September 8
From 2008.igem.org
(Difference between revisions)
(→Condition tested) |
(→Condition tested) |
||
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Calendar_Links|September 7|September 9}} | {{Paris/Calendar_Links|September 7|September 9}} | ||
| + | =Constructions= | ||
| + | ==Results of the transformation, we did yesterday== | ||
| + | {|border="1" style="text-align: center" | ||
| + | |'''Ligation n°''' | ||
| + | |'''Insert''' | ||
| + | |'''Vector''' | ||
| + | |'''Construction''' | ||
| + | |'''Antibio''' | ||
| + | |'''Number of colonies''' | ||
| + | |- | ||
| + | |positive control | ||
| + | | | ||
| + | | | ||
| + | |puc 19 | ||
| + | |Amp | ||
| + | | +++ | ||
| + | |- | ||
| + | |L173 | ||
| + | |D112 | ||
| + | |D187 | ||
| + | |rbs-tetR-mRFP-LVA+ | ||
| + | |Amp | ||
| + | |0 | ||
| + | |- | ||
| + | |L183 | ||
| + | |D134 | ||
| + | |D187 | ||
| + | | pFlhB - mRFP LVA+ | ||
| + | |Amp | ||
| + | |0 | ||
| + | |- | ||
| + | |L184 | ||
| + | |D149 | ||
| + | |D198 | ||
| + | | pFliL - ECFP LVA+ | ||
| + | |Amp | ||
| + | |0 | ||
| + | |- | ||
| + | |L185 | ||
| + | |D149 | ||
| + | |D199 | ||
| + | | pFliL - ECFP LVA- | ||
| + | |Amp | ||
| + | |0 | ||
| + | |} | ||
| + | |||
| + | =>This is a supplementary time that the experiment of ligation-transformation don't work.<br> It means that on of this step have a mistake. <br> So we need to do several test to understand where the problem is located.<br> We Hypothesis that the problem is due to a defectious Ligase. | ||
| + | |||
=Checking our ligases= | =Checking our ligases= | ||
Since our ligation experiments didn't work during these few days, we presume that our problem came from the ligases. That's why we decided to check the activity of the ligases in our possession. | Since our ligation experiments didn't work during these few days, we presume that our problem came from the ligases. That's why we decided to check the activity of the ligases in our possession. | ||
| Line 5: | Line 53: | ||
* we will digest a plasmid with only one restriction enzyme (EcoRI). | * we will digest a plasmid with only one restriction enzyme (EcoRI). | ||
* then we will try to ligate it back using our ligases. | * then we will try to ligate it back using our ligases. | ||
| + | |||
==Condition tested == | ==Condition tested == | ||
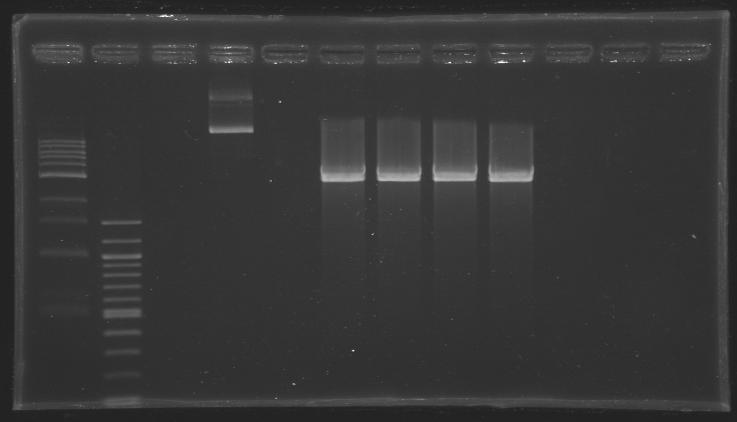
| - | [[Image:KR000307.JPG|thumb|Before gel excision | + | [[Image:KR000307.JPG|thumb|Before gel excision <br> well n°4: undigested plasmid (MP101.1) <br> well °6, 7, 8, 9: digested plasmid (D202)]] |
| - | + | ||
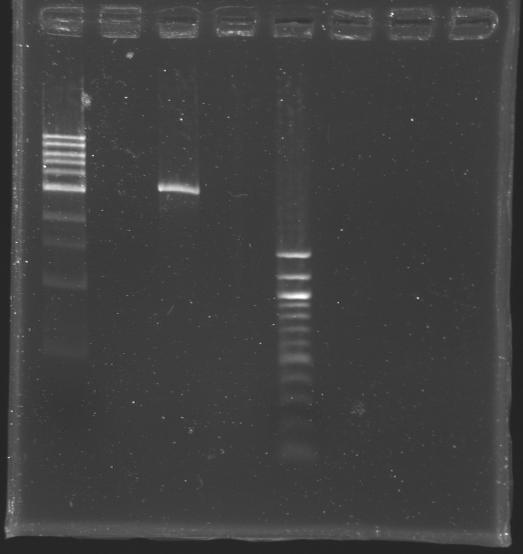
[[Image:KR000309.JPG|thumb|After purification]] | [[Image:KR000309.JPG|thumb|After purification]] | ||
| Line 51: | Line 99: | ||
==Digestion== | ==Digestion== | ||
| - | + | Reaction mixture | |
(carried out four times ) | (carried out four times ) | ||
| - | * ADN (MP101.1) | + | * ADN (MP101.1): 3.8 µL |
| - | * H2O 21.9 µL | + | * H2O: 21.9 µL |
| - | * 10X Buffer 3 µL | + | * 10X Buffer: 3 µL |
| - | * BSA 0.3 µl | + | * BSA: 0.3 µl |
| - | * EcoRI 1 µL | + | * EcoRI: 1 µL |
==Purification== | ==Purification== | ||
*We purifed our digestion products by gel extraction (Qiagen kit) | *We purifed our digestion products by gel extraction (Qiagen kit) | ||
| - | *we obtained | + | *After elution, we obtained [DNA] ~ 25 ng/µL (based on the intensity of the band) |
== Overnight ligation (16°C)== | == Overnight ligation (16°C)== | ||
| - | * DNA 4 µL | + | * DNA: 4 µL |
| - | * 10X | + | * 10X Buffer: 2 µL |
| - | * | + | * Ligase: 1 µL |
| - | * H2O 13 µL | + | * H2O: 13 µL |
Latest revision as of 19:31, 16 September 2008
ConstructionsResults of the transformation, we did yesterday
=>This is a supplementary time that the experiment of ligation-transformation don't work. Checking our ligasesSince our ligation experiments didn't work during these few days, we presume that our problem came from the ligases. That's why we decided to check the activity of the ligases in our possession. To do so:
Condition tested
DigestionReaction mixture (carried out four times )
Purification
Overnight ligation (16°C)
|
 "
"