Team:Paris/August 11
From 2008.igem.org
(→Culture of ligation transformants) |
(→Culture of ligation transformants) |
||
| (54 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Calendar_Links|August 10|August 12}} | {{Paris/Calendar_Links|August 10|August 12}} | ||
| - | == | + | ==Transformation== |
| + | All the ligations were transformed according [[Team:Paris/Notebook/Protocols#Transformation|transformation for Top10 protocol]] | ||
| - | *4 clones of each transformation were cultured in '''7,5 mL LB + ampicilline'''. The colonies picked up were the rest of those already picked up from the transformation plate (for PCR screening). | + | |
| + | |||
| + | ==PCR== | ||
| + | |||
| + | ''We performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.'' | ||
| + | |||
| + | |||
| + | === '''PCR amplification''' === | ||
| + | |||
| + | ==== '''Protocol''' ==== | ||
| + | |||
| + | * '''List of Oligos''' : | ||
| + | {| | ||
| + | |- style="background: #649CD7;" | ||
| + | |- style="background: #649CD7; text-align: center;" | ||
| + | |width=5%| Number | ||
| + | |width=15%| Name | ||
| + | |width=40%| Sequence | ||
| + | |width=5%| Length | ||
| + | |width=30%| Comments | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O110 | ||
| + | | FlhDC-Uri-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGTTGTATGTGCGTGTAGTGACGAGTACAG | ||
| + | | 58 | ||
| + | |Don't amplify the both OmpR binding site | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O111 | ||
| + | | FlhDC-Total-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGTCATTTTTGCTTGCTAGCGTACGGAAAA | ||
| + | | 58 | ||
| + | |Amplify the both OmpR binding site | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O113 | ||
| + | | FlhDC(nu)-R | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTACAGAATAACCAACTTTATTTTTATG | ||
| + | | 54 | ||
| + | |Don't amplify the natural rbs of FlhD (only promoter) | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O124 | ||
| + | | Oligo-pfliL-Forward-TRO | ||
| + | | TCGAATTCGCGGCCGCTTCTAGAGCAAGGGCGTGTAACAGGCAAC | ||
| + | | 45 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O125 | ||
| + | | Oligo-pfliL-Reverse-TRO | ||
| + | | TCCTGCAGCGGCCGCTACTAGTAGTCATGTGTTGCGGTCTTCCTGTG | ||
| + | | 47 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O130 | ||
| + | | Gene-FlhC-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGATGAGTGAAAAAAGCATTGTTCAGG | ||
| + | | 53 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O131 | ||
| + | | Gene-FlhC-R-TRO | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTAAACAGCCTGTACTCTCTGTTCATCC | ||
| + | | 60 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O132 | ||
| + | | Gene-FlhD-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGATGCATACCTCCGAGTTGCTGAAAC | ||
| + | | 53 | ||
| + | |Don't amplify the natural rbs of FlhD | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O133 | ||
| + | | Gene-FlhD-R-TRO | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTAGGCCCTTTTCTTGCGCAGCGCTTCT | ||
| + | | 60 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O140 | ||
| + | | PlasTest_GFP_Rv | ||
| + | | CCCTGCAGTTAATTAATATAAACGCAGAAAGGCCAC | ||
| + | | 36 | ||
| + | |Amplify E0240 with a PacI and a PstI restriction site at the end | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O141 | ||
| + | | PTst_GFPrbs+_F | ||
| + | | GCCTGCAGTCACACAGGAAAGTACTAGATGC | ||
| + | | 31 | ||
| + | |Amplify E0240 with the promoter and a PstI restriction site at the beginning | ||
| + | |} | ||
| + | |||
| + | |||
| + | * '''Preparation of the templates''' : <br> Resuspension of 1 colony in 100µl of water. | ||
| + | |||
| + | |||
| + | * '''Preparation of PCR mix''' : | ||
| + | ''For each sample,'' | ||
| + | |||
| + | 1 µl dNTP | ||
| + | <br>10 µl Buffer Phusion 5x | ||
| + | <br>2,5 µl Oligo_F | ||
| + | <br>2,5 µl Oligo_R | ||
| + | <br>1µl template | ||
| + | <br>1 µl Phusion | ||
| + | <br>50 µl qsp H2O (33µl) | ||
| + | |||
| + | |||
| + | {| border="1" | ||
| + | |- style="text-align: center;" | ||
| + | |'''Name''' | ||
| + | |'''genes''' | ||
| + | |'''Oligo''' | ||
| + | |'''templates''' | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_130 | ||
| + | |E0240 RBS+ | ||
| + | |O141_O140 | ||
| + | |MP143 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_130' | ||
| + | | - | ||
| + | |O141_O140 | ||
| + | |Water | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_131 | ||
| + | |flhD RBS- | ||
| + | |O132_O133 | ||
| + | |MG1655 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_131' | ||
| + | | - | ||
| + | |O132_O133 | ||
| + | |Water | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_132 | ||
| + | |flhC RBS- | ||
| + | |O130_O131 | ||
| + | |MG1655 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_132' | ||
| + | | - | ||
| + | |O130_O131 | ||
| + | |Water | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_133 | ||
| + | |flhDC + prom | ||
| + | |O110_O131 | ||
| + | |MG1655 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_133' | ||
| + | | - | ||
| + | |O110_O131 | ||
| + | |Water | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_134 | ||
| + | |flhDC + prom | ||
| + | |O111_O131 | ||
| + | |MG1655 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_134' | ||
| + | | - | ||
| + | |O111_O131 | ||
| + | |Water | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_135 | ||
| + | |pfliL | ||
| + | |O124_O125 | ||
| + | |MG1655 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_135' | ||
| + | | - | ||
| + | |O124_O125 | ||
| + | |Water | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_136 | ||
| + | |pflhDC | ||
| + | |O110_O113 | ||
| + | |MG1655 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_136' | ||
| + | | - | ||
| + | |O110_O113 | ||
| + | |Water | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_137 | ||
| + | |pflhDC | ||
| + | |O111_O113 | ||
| + | |MG1655 | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_137' | ||
| + | | - | ||
| + | |O111_O113 | ||
| + | |Water | ||
| + | |} | ||
| + | |||
| + | |||
| + | * Program PCR_Screening : Annealing 30" at 60°C - Time élongation 1'30" at 72°C - Number cycle : 30 | ||
| + | |||
| + | ==== '''PCR verification/Analysis''' ==== | ||
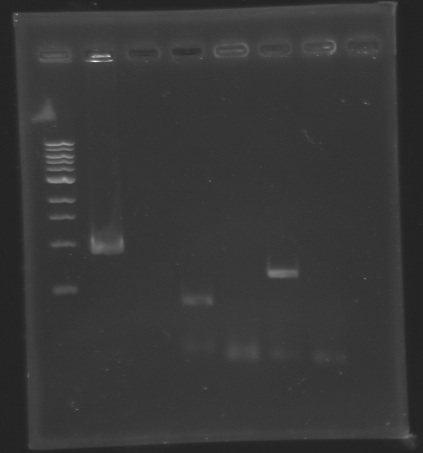
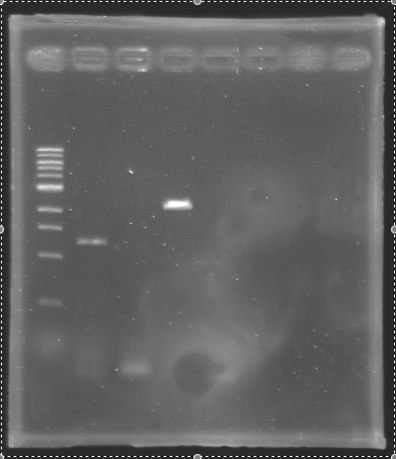
| + | [[Image:KR000148.jpg|thumb|Analysis of PCR product (Gel 1)]] [[Image:KR000147bis.jpg|thumb|Analysis of PCR product (Gel 2)]] | ||
| + | ''After the PCR :'' | ||
| + | * 2*3µl have been analysed by electrophoresis | ||
| + | * the other 44µl of PCR products have been purified by the Promega kit. | ||
| + | |||
| + | |||
| + | * '''Electrophoresis''' | ||
| + | |||
| + | ladder : 10µl ladder 1 kb | ||
| + | <br> samples : 3µl of PCR products + 2µl of Loading Dye | ||
| + | <br> migration 30min at 100V, on a '''1%''' agarose gel | ||
| + | |||
| + | |||
| + | * '''Results :''' | ||
| + | {| border="1" | ||
| + | |- style="text-align: center;" | ||
| + | |'''Name''' | ||
| + | |'''Promotor''' | ||
| + | |align="center"|'''Gel''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_130 | ||
| + | |E0240 | ||
| + | |Gel 1 | ||
| + | |2 | ||
| + | |876 bp | ||
| + | |style="background: #cbff7B"|<center> 900 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_130' | ||
| + | |Negative Control | ||
| + | |Gel 1 | ||
| + | |3 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_131 | ||
| + | |flhD RBS- | ||
| + | |Gel 1 | ||
| + | |4 | ||
| + | |351 bp | ||
| + | |style="background: #cbff7B"|<center> 350 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_131' | ||
| + | |Negative Control | ||
| + | |Gel 1 | ||
| + | |5 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_132 | ||
| + | |flhC RBS- | ||
| + | |Gel 1 | ||
| + | |6 | ||
| + | |579 bp | ||
| + | |style="background: #cbff7B"|<center> 600 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_132' | ||
| + | |Negative Control | ||
| + | |Gel 1 | ||
| + | |7 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_133 | ||
| + | |flhDC + prom | ||
| + | |Gel 2 | ||
| + | |2 | ||
| + | |1165 bp | ||
| + | |style="background: #cbff7B"|<center> 1300 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_133' | ||
| + | |Negative Control | ||
| + | |Gel 2 | ||
| + | |3 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_134 | ||
| + | |flhDC + prom | ||
| + | |Gel 2 | ||
| + | |4 | ||
| + | |1311 bp | ||
| + | |style="background: #ff6d73"|<center> 2100 pb </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_134' | ||
| + | |Negative Control | ||
| + | |Gel 2 | ||
| + | |5 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 pb</center> | ||
| + | |} | ||
| + | |||
| + | |||
| + | ==> '''Conclusion :''' We observed the size expected for the PCR products, but not for pflhDC (PCR_134), is right. We hypothesis for PCR_138 that the size is longer that expected due to the aspecific fixation of Oligo O111 (upper to the real site). | ||
| + | |||
| + | |||
| + | * '''Electrophoresis''' | ||
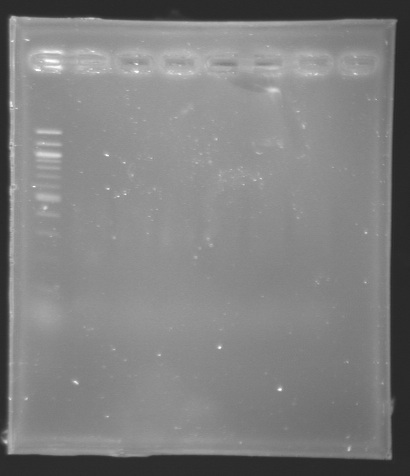
| + | [[Image:KR000150.jpg|thumb|Analysis of PCR product (Gel 3)]] | ||
| + | ladder : 10µl ladder 100 bp | ||
| + | <br> samples : 3µl of PCR products + 2µl of Loading Dye | ||
| + | <br> migration 30min at 100V, on a '''2%''' agarose gel | ||
| + | |||
| + | |||
| + | * '''Results :''' | ||
| + | {| border="1" | ||
| + | |- style="text-align: center;" | ||
| + | |'''Name''' | ||
| + | |'''Promotor''' | ||
| + | |align="center"|'''Gel''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_135 | ||
| + | |pfliL | ||
| + | |Gel 3 | ||
| + | |2 | ||
| + | |124 bp | ||
| + | |style="background: #ff6d73"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_135' | ||
| + | |Negative Control | ||
| + | |Gel 3 | ||
| + | |3 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_136 | ||
| + | |pflhDC | ||
| + | |Gel 3 | ||
| + | |4 | ||
| + | |223 | ||
| + | |style="background: #ff6d73"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_136' | ||
| + | |Negative Control | ||
| + | |Gel 3 | ||
| + | |5 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_137 | ||
| + | |pflhDC | ||
| + | |Gel 3 | ||
| + | |6 | ||
| + | |369 | ||
| + | |style="background: #ff6d73"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_137' | ||
| + | |Negative Control | ||
| + | |Gel 3 | ||
| + | |7 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |} | ||
| + | |||
| + | |||
| + | ==> '''Conclusion :''' We need to repeat the experiments. | ||
| + | |||
| + | ==Culture of ligation transformants (pFlgA, pFlgB and pFlhB)== | ||
| + | |||
| + | *4 clones of each transformation were cultured in '''7,5 mL LB + ampicilline'''. The colonies picked up were the rest of those already picked up from the transformation plate (for the PCR screening of August 8). | ||
*37°C overnight | *37°C overnight | ||
{|| Border="1" | {|| Border="1" | ||
| - | | | + | |- style="text-align: center;" |
| - | | colspan="4 | + | |'''Ligation''' |
| - | | colspan="4 | + | | colspan="4"|L128 |
| - | | colspan="4 | + | | colspan="4"|L129 |
| - | |- | + | | colspan="4"|L130 |
| - | + | |- style="text-align: center;" | |
| - | | colspan="4 | + | |'''Name''' |
| - | | colspan="4 | + | | colspan="4"| pFlgA |
| - | | colspan="4 | + | | colspan="4"| pFlgB |
| - | |- | + | | colspan="4"|pFlhB |
| - | + | |- style="text-align: center;" | |
| - | + | |'''Clone N°''' | |
| - | + | |1 | |
| - | + | |2 | |
| - | + | |3 | |
| - | + | |4 | |
| - | + | |1 | |
| - | + | |2 | |
| - | + | |6 | |
| - | + | |7 | |
| - | + | |1 | |
| - | + | |2 | |
| - | + | |7 | |
| - | |- | + | |8 |
| - | + | |- style="text-align: center;" | |
| - | | | + | |'''Red fluorescence''' |
| - | | | + | |style="background: #ff6d73"|yes |
| - | + | |style="background: #ff6d73"|yes | |
| - | + | |no | |
| - | | | + | |no |
| - | | | + | |style="background: #ff6d73"|yes |
| - | + | |style="background: #ff6d73"|yes | |
| - | + | |no | |
| - | | | + | |no |
| - | + | |style="background: #ff6d73"|yes | |
| - | + | |no | |
| - | + | |no | |
| + | |no | ||
|} | |} | ||
Latest revision as of 16:11, 18 August 2008
TransformationAll the ligations were transformed according transformation for Top10 protocol
PCRWe performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR amplificationProtocol
For each sample, 1 µl dNTP
PCR verification/AnalysisAfter the PCR :
ladder : 10µl ladder 1 kb
ladder : 10µl ladder 100 bp
Culture of ligation transformants (pFlgA, pFlgB and pFlhB)
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"