Team:Paris/August 13
From 2008.igem.org
(Difference between revisions)
(→Sequencing Minipreps we did yesterday) |
(→PCR screening) |
||
| (43 intermediate revisions not shown) | |||
| Line 68: | Line 68: | ||
|'''Name''' | |'''Name''' | ||
|'''Ligation''' | |'''Ligation''' | ||
| - | |||
|'''Description''' | |'''Description''' | ||
| + | |'''Biobricks''' | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
|MP147.1 | |MP147.1 | ||
|L100.1 | |L100.1 | ||
| rowspan="3"| rbs TetR - ECFP<br>D110 (BV) - D130 (BI) | | rowspan="3"| rbs TetR - ECFP<br>D110 (BV) - D130 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|MP147.2 | |MP147.2 | ||
| Line 85: | Line 85: | ||
|L101.1 | |L101.1 | ||
| rowspan="3"| rbs TetR - GFP tripart<br>D110 (BV) - D131 (BI) | | rowspan="3"| rbs TetR - GFP tripart<br>D110 (BV) - D131 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|MP148.2 | |MP148.2 | ||
| Line 96: | Line 96: | ||
|L114.1 | |L114.1 | ||
| rowspan="3"| AracpBAD - gfp tripart<br>D126 (BV) - D131 (BI) | | rowspan="3"| AracpBAD - gfp tripart<br>D126 (BV) - D131 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_regulatory.png]][[Image:Part_icon_reporter.png]] | + | | rowspan="3"| [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|MP149.2 | |MP149.2 | ||
| Line 107: | Line 107: | ||
|L120.1 | |L120.1 | ||
| rowspan="3"| tetR repressible promoter - ECFP<br>D106 (BV) - D130 (BI) | | rowspan="3"| tetR repressible promoter - ECFP<br>D106 (BV) - D130 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|MP150.2 | |MP150.2 | ||
| Line 118: | Line 118: | ||
|L122.1 | |L122.1 | ||
| RBS-lasI - ECFP <br>D107 (BV) - D130 (BI) | | RBS-lasI - ECFP <br>D107 (BV) - D130 (BI) | ||
| - | | [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|MP152.1 | |MP152.1 | ||
|L123.1 | |L123.1 | ||
| - | | rowspan="3"| RBS lasI - | + | | rowspan="3"| RBS lasI - gfp tripart<br>D107 (BV) - D131 (BI) |
| - | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|MP151.2 | |MP151.2 | ||
| Line 144: | Line 144: | ||
|MP154.1 | |MP154.1 | ||
|L132.1 | |L132.1 | ||
| - | | rowspan="3"| | + | | rowspan="3"|flhDC <br> D142(FV) - D139(FI) |
| - | | rowspan="3"| | + | | rowspan="3"|[[Image:Icon_coding.png]] |
|- | |- | ||
|MP154.2 | |MP154.2 | ||
| Line 155: | Line 155: | ||
|MP155.1 | |MP155.1 | ||
|L133.1 | |L133.1 | ||
| - | | rowspan="1"| | + | | rowspan="1"|OmpR*<br> D142(FV) - D140(FI) |
| - | | rowspan="1"| | + | | rowspan="1"|[[Image:Icon_coding.png]] |
|- | |- | ||
|MP156.1 | |MP156.1 | ||
|L134.1 | |L134.1 | ||
| - | | rowspan="3"| | + | | rowspan="3"|EnvZ*<br> D142(FV) - D141(FI) |
| - | | rowspan="3"| | + | | rowspan="3"|[[Image:Icon_coding.png]] |
|- | |- | ||
|MP156.2 | |MP156.2 | ||
| Line 171: | Line 171: | ||
|MP157.1 | |MP157.1 | ||
|L138.1 | |L138.1 | ||
| - | | rowspan="1"| | + | | rowspan="1"|gfp generator (E0240)<br> D137(FV) - D138(FI) |
| - | | rowspan="1"| | + | | rowspan="1"|[[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|} | |} | ||
| Line 189: | Line 189: | ||
|L100.1 | |L100.1 | ||
| rowspan="3"| rbs TetR - ECFP<br>D110 (BV) - D130 (BI) | | rowspan="3"| rbs TetR - ECFP<br>D110 (BV) - D130 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|S146.2 | |S146.2 | ||
| Line 200: | Line 200: | ||
|L101.1 | |L101.1 | ||
| rowspan="3"| rbs TetR - GFP tripart<br>D110 (BV) - D131 (BI) | | rowspan="3"| rbs TetR - GFP tripart<br>D110 (BV) - D131 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|S147.2 | |S147.2 | ||
| Line 222: | Line 222: | ||
|L120.1 | |L120.1 | ||
| rowspan="3"| tetR repressible promoter - ECFP<br>D106 (BV) - D130 (BI) | | rowspan="3"| tetR repressible promoter - ECFP<br>D106 (BV) - D130 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|S149.2 | |S149.2 | ||
| Line 233: | Line 233: | ||
|L122.1 | |L122.1 | ||
| RBS-lasI - ECFP <br>D107 (BV) - D130 (BI) | | RBS-lasI - ECFP <br>D107 (BV) - D130 (BI) | ||
| - | | [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
|S151.1 | |S151.1 | ||
|L123.1 | |L123.1 | ||
| rowspan="3"| RBS lasI - ECFP<br>D107 (BV) - D131 (BI) | | rowspan="3"| RBS lasI - ECFP<br>D107 (BV) - D131 (BI) | ||
| - | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]] | + | | rowspan="3"| [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] |
|- | |- | ||
| - | | | + | |S151.2 |
|L123.2 | |L123.2 | ||
|- | |- | ||
| - | | | + | |S151.3 |
|L123.3 | |L123.3 | ||
|- | |- | ||
| Line 250: | Line 250: | ||
| rowspan="3"| Strongest RBS (1)- LasR activator (+LVA)<br>D102 (BV) - D114 (BI) | | rowspan="3"| Strongest RBS (1)- LasR activator (+LVA)<br>D102 (BV) - D114 (BI) | ||
| rowspan="3"|[[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]] | | rowspan="3"|[[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]] | ||
| + | |- | ||
| + | |S152.2 | ||
| + | |L126.2 | ||
| + | |- | ||
| + | |S152.3 | ||
| + | |L126.3 | ||
|} | |} | ||
| - | ==PCR screening== | + | ==Assay to purify a PCR product using the Qiaquick Gel Extraction kit== |
| + | |||
| + | *sample used: yesterday PCR product of L130.8 (~40 µL) | ||
| + | *protocol used: Qiagen PCR purification kit | ||
| + | *use of buffer QG (Gel Extraction kit) instead of buffer PBI (PCR purification kit) | ||
| + | *elution in 30 µL of buffer EB | ||
| + | *30 µL of purified product + 12 µL of loading blue | ||
| + | *electrophoresis, 10 µL loaded | ||
| + | => see '''Gel 1''', well n° 8 | ||
| + | |||
| + | ==> results: PCR products can be purified using the Qiaquick Gel Extraction kit. We just have to replace the buffer PBI by the buffer QG! | ||
| + | |||
| + | == '''PCR screening'''== | ||
| + | |||
| + | === Electrophoresis Setting === | ||
4 more transformants of L130 (pFlhB into J61002) are screened by PCR. | 4 more transformants of L130 (pFlhB into J61002) are screened by PCR. | ||
| - | *PCR screening programm | + | *PCR screening programm; elogation time: 1 min 30 |
| - | + | ||
*template: colonies from the transformation plate | *template: colonies from the transformation plate | ||
*positive control: S142 (J61002) | *positive control: S142 (J61002) | ||
| Line 262: | Line 281: | ||
| + | ===Results of Electrophoresis=== | ||
| + | |||
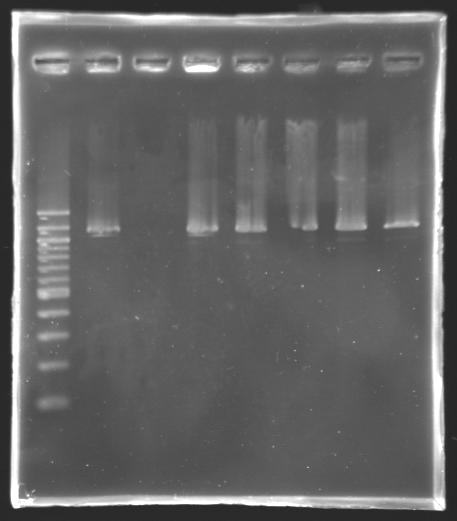
| + | [[Image:Screening-PCR-080813.JPG|thumb|'''Gel 1''': PCR screening ('''2-7''') & PCR product purification using the Qiaquick Gel Extraction kit ('''8''')]] | ||
| + | |||
| + | *1% agarose gel | ||
| + | *10 µL loaded | ||
| + | |||
| + | {| style="text-align: center;" border="1" | ||
| + | |'''well n°''' | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |7 | ||
| + | |8 | ||
| + | |- | ||
| + | |'''sample''' | ||
| + | |100 bp DNA ladder | ||
| + | |positive control | ||
| + | |negative control | ||
| + | |'''L130.3''' | ||
| + | |'''L130.4''' | ||
| + | |'''L130.5''' | ||
| + | |'''L130.6''' | ||
| + | |PCR product (L130.8) purified by the Qiaquick Gel Extraction kit | ||
| + | |- | ||
| + | |'''red fluorescence''' | ||
| + | |rowspan="3"| | ||
| + | |strain a little bit pink | ||
| + | |not concerned | ||
| + | |no | ||
| + | |no | ||
| + | |no | ||
| + | |no | ||
| + | |rowspan="3"|not concerned | ||
| + | |- | ||
| + | |'''expected size''' | ||
| + | |1,161 kb | ||
| + | |0 kb | ||
| + | |colspan="4"|<center>'''1,338 kb'''</center> | ||
| + | |- | ||
| + | |'''measured size''' | ||
| + | |style="background: #cbff7B"| 1,2 kb | ||
| + | |style="background: #cbff7B"| 0 kb | ||
| + | |style="background: #ff6d73"| 1,2 kb | ||
| + | |style="background: #ff6d73"| 1,2 kb | ||
| + | |style="background: #ff6d73"| 1,2 kb | ||
| + | |style="background: #ff6d73"| 1,2 kb | ||
| + | |} | ||
| + | ==> The L130 transformants analysed are not correct. | ||
| - | == | + | =='''Transformation of the ligation we did [[Team:Paris/August_12|yesterday]]'''== |
| + | We transformed L 139, L140, L141 and L142 following the [[Team:Paris/Notebook/Protocols#Transformation|standard protocol using Invitrogen's TOP 10 chemically competent cells]]. The positive control is a transformation with pUC19 and the negative control has no plasmid. | ||
Latest revision as of 16:12, 15 August 2008
Sequencing Minipreps we did yesterday
Minipreps: Plasmid extraction
Glycerol Stocks
Assay to purify a PCR product using the Qiaquick Gel Extraction kit
=> see Gel 1, well n° 8 ==> results: PCR products can be purified using the Qiaquick Gel Extraction kit. We just have to replace the buffer PBI by the buffer QG! PCR screeningElectrophoresis Setting4 more transformants of L130 (pFlhB into J61002) are screened by PCR.
Results of Electrophoresis
==> The L130 transformants analysed are not correct. Transformation of the ligation we did yesterdayWe transformed L 139, L140, L141 and L142 following the standard protocol using Invitrogen's TOP 10 chemically competent cells. The positive control is a transformation with pUC19 and the negative control has no plasmid. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"