Team:Paris/August 25
From 2008.igem.org
(Difference between revisions)
AnaJimenez (Talk | contribs) (→Promoter characterization plasmids) |
AnaJimenez (Talk | contribs) (→PCR screenning: transformation results from August 23th) |
||
| (27 intermediate revisions not shown) | |||
| Line 38: | Line 38: | ||
Aim : Construction of ''' "pFlgA-RBS-GFP-dbl ter" (pFlgA-E0240)''' [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | Aim : Construction of ''' "pFlgA-RBS-GFP-dbl ter" (pFlgA-E0240)''' [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| - | |||
=='''Results of the transformation we did [[Team:Paris/August 23 |the day before yesterday]]'''== | =='''Results of the transformation we did [[Team:Paris/August 23 |the day before yesterday]]'''== | ||
| Line 84: | Line 83: | ||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| + | |'''digestion number''' | ||
|'''name''' | |'''name''' | ||
| - | |''' | + | |'''template''' |
| - | |''' | + | |'''Enzymes''' |
| - | + | ||
|- | |- | ||
| + | |D159 | ||
|EnvZ* | |EnvZ* | ||
|PCR129 from August 8th | |PCR129 from August 8th | ||
|XbaI & PstI | |XbaI & PstI | ||
| - | |||
|- | |- | ||
| + | |D116 | ||
|pSB1A2 | |pSB1A2 | ||
|MP108 (C0179 (lasR-pSB1A2)) | |MP108 (C0179 (lasR-pSB1A2)) | ||
|XbaI & PstI | |XbaI & PstI | ||
| - | |||
|- | |- | ||
|} | |} | ||
| Line 113: | Line 112: | ||
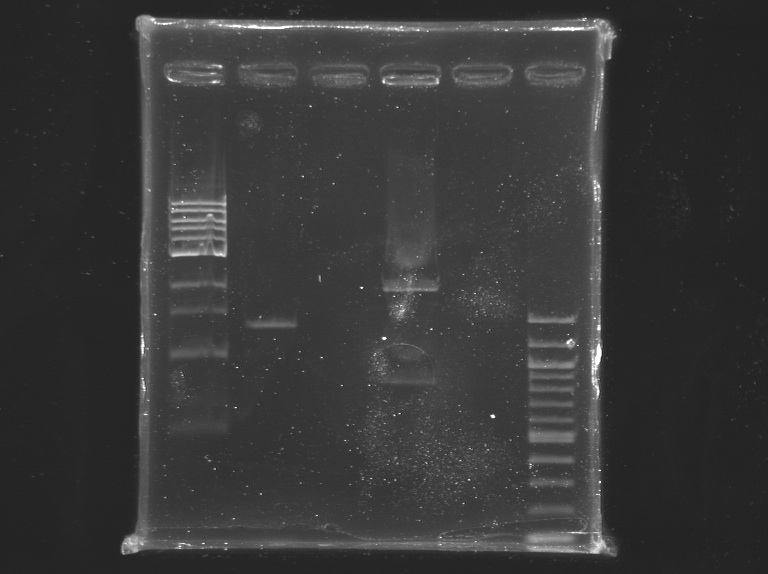
[[Image:KR000225.jpg|thumb|]] | [[Image:KR000225.jpg|thumb|]] | ||
| - | |||
1% agarose gel | 1% agarose gel | ||
| Line 162: | Line 160: | ||
| + | ='''Screening of the cloning of pFlgA-YFP Tripart (LVA+/-)'''= | ||
| - | = | + | ==Electrophoresis== |
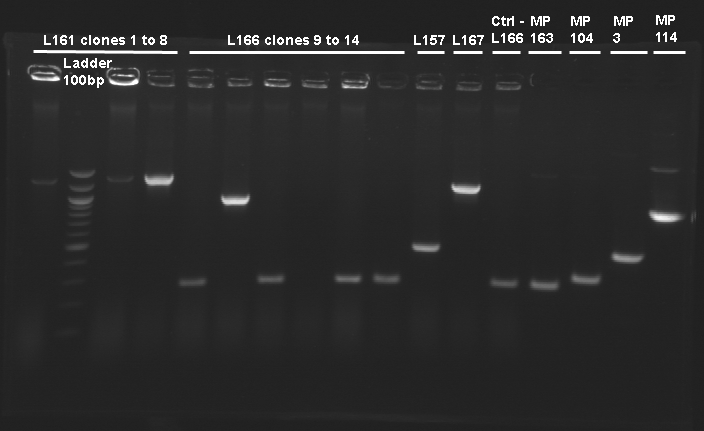
| + | [[Image:KR000232.JPG|thumb|Gel n°1 : Screening of L160]] | ||
| + | [[Image:KR000229.JPG|thumb|Gel n°2 : Screening of L161]] | ||
| + | {|border="1" style="text-align: center" | ||
| + | | | ||
| + | |colspan="16"|Gel n° 1 | ||
| + | |- | ||
| + | |'''well n°''' | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |7 | ||
| + | |8 | ||
| + | |9 | ||
| + | |10 | ||
| + | |11 | ||
| + | |12 | ||
| + | |13 | ||
| + | |14 | ||
| + | |15 | ||
| + | |16 | ||
| + | |- | ||
| + | |'''sample''' | ||
| + | |100pb ladder | ||
| + | |colspan="7"|don't matter | ||
| + | |L160.1 | ||
| + | |L160.2 | ||
| + | |L160.3 | ||
| + | |L160.4 | ||
| + | |L160.5 | ||
| + | |L160.6 | ||
| + | |L160.7 | ||
| + | |L160.8 | ||
| + | |- | ||
| + | |'''expected size (pb)''' | ||
| + | | | ||
| + | |colspan="7"| | ||
| + | |colspan="8"|1 200 | ||
| + | |- | ||
| + | |'''measured size (pb)''' | ||
| + | | | ||
| + | |colspan="7"| | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |style="background: #cbff7B"|1 200 | ||
| + | |} | ||
| - | == | + | {|border="1" style="text-align: center" |
| + | | | ||
| + | |colspan="16"|Gel n° 2 | ||
| + | |- | ||
| + | |'''well n°''' | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |7 | ||
| + | |8 | ||
| + | |9 | ||
| + | |10 | ||
| + | |11 | ||
| + | |12 | ||
| + | |13 | ||
| + | |14 | ||
| + | |15 | ||
| + | |16 | ||
| + | |- | ||
| + | |'''sample''' | ||
| + | |L161.1 | ||
| + | |100 pb ladder | ||
| + | |L161.2 | ||
| + | |L161.3 | ||
| + | |colspan="12"|don't matter | ||
| + | |- | ||
| + | |'''expected size (pb)''' | ||
| + | |1 167 | ||
| + | | | ||
| + | |colspan="2"|1 167 | ||
| + | |- | ||
| + | |'''measured size (pb)''' | ||
| + | |style="background: #cbff7B"|1 300 | ||
| + | | | ||
| + | |style="background: #cbff7B"|1 300 | ||
| + | |style="background: #cbff7B"|1 30 | ||
| + | |} | ||
| - | + | ==Minipreps and glycerol stock== | |
| + | |||
| + | {|border="1" style="text-align: center" | ||
| + | |'''Miniprep''' | ||
| + | |'''Glycerol Stock | ||
| + | |'''Ligation''' | ||
| + | |'''Name''' | ||
| + | |- | ||
| + | |MP1 | ||
| + | |S1 | ||
| + | |L160 | ||
| + | |FlgA-rbs-YFP-dbl ter | ||
| + | |- | ||
| + | |MP1 | ||
| + | |S1 | ||
| + | |L161 | ||
| + | |FlgA-rbs-YFP-LVA+-dbl ter | ||
| + | |} | ||
| + | |||
| + | |||
| + | ='''Promoter characterization plasmids'''= | ||
| + | |||
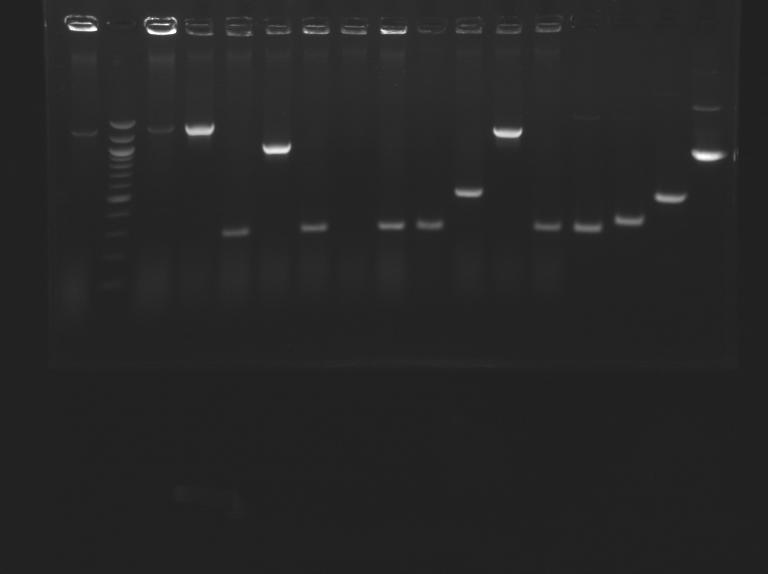
| + | ==PCR screenning: transformation results from August 23th== | ||
[[Team:Paris/Notebook/Protocols#Transformation |Protocol]] | [[Team:Paris/Notebook/Protocols#Transformation |Protocol]] | ||
| + | |||
| + | [[Image:25-08-08.png|200px]] | ||
| + | [[Image:25-08-08bis.png|200px]] | ||
| + | |||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
|'''Ligation name''' | |'''Ligation name''' | ||
| - | |''' | + | |'''Clone number''' |
| - | + | ||
| - | + | ||
| - | + | ||
|'''Product description''' | |'''Product description''' | ||
| - | |''' | + | |'''Primers used''' |
| - | |''' | + | |'''Size expected''' |
| + | |'''Size observed''' | ||
|- | |- | ||
| - | | | + | |no name ligation |
| - | | | + | |1 |
| - | | | + | |tetR-B0015 |
| - | + | |O18-O19 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |- | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| | | | ||
| - | |||
| | | | ||
| - | |||
| - | |||
| - | |||
|- | |- | ||
| - | | | + | |Control L166 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|1 | |1 | ||
| + | |Vector autoligation control | ||
| + | |O18-O19 | ||
| + | |249 | ||
| + | |correct | ||
|- | |- | ||
| - | | | + | |L166 |
| - | | | + | |1-5,7-9,11-14 |
| + | |RBS B0032 - tetR | ||
| + | |O18-O19 | ||
| | | | ||
| - | | | + | |around 249 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
|L166 | |L166 | ||
| - | | | + | |6 |
| - | + | ||
| - | + | ||
| - | + | ||
|RBS B0032 - tetR | |RBS B0032 - tetR | ||
| - | | | + | |O18-O19 |
| - | | | + | | |
| + | | | ||
|- | |- | ||
| - | | | + | |L166 |
| - | | | + | |10 |
| + | |RBS B0032 - tetR | ||
| + | |O18-O19 | ||
| | | | ||
| - | |||
| | | | ||
| - | |||
| - | |||
| - | |||
|- | |- | ||
|L167 | |L167 | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|1 | |1 | ||
| + | |gfp generator - pTet | ||
| + | |O18-O19 | ||
| + | | | ||
| + | | | ||
|} | |} | ||
Latest revision as of 19:27, 9 September 2008
Construction for SynchronizationTransformation of the ligations we did yesterday
Construction of pFlgA - GFP GeneratorAim : Construction of "pFlgA-RBS-GFP-dbl ter" (pFlgA-E0240) Results of the transformation we did the day before yesterday
=> Need to screen to know which clones we can use for the of pFlgA promotor characterization. Cloning of EnvZ*The sequencing of EnvZ* previously cloned, revealed a loss of about 300 bp. EnvZ* contains indeed an EcoRI restriction site within its sequence. So we can't use this enzyme during the cloning. Digestion
Reaction mixture
Incubation at 37°C during 2H25, and then ~20 min at 65°C Electrophoresis1% agarose gel
elution in 30 µL of buffer EB
Screening of the cloning of pFlgA-YFP Tripart (LVA+/-)Electrophoresis
Minipreps and glycerol stock
Promoter characterization plasmidsPCR screenning: transformation results from August 23th
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"