Team:Paris/August 27
From 2008.igem.org
(Difference between revisions)
(→Transformation results) |
|||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Calendar_Links|August 26|August 28}} | {{Paris/Calendar_Links|August 26|August 28}} | ||
| - | = | + | =Construction of pFlhB - mRFP Tripart (LVA+)= |
Aim : Construction of ''' "pFlhB-RBS-mRFP-dbl ter" (pFlhB-I732078)''' [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | Aim : Construction of ''' "pFlhB-RBS-mRFP-dbl ter" (pFlhB-I732078)''' [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| - | We can only do the construction with mRFP Tripart (LVA+) because the stable strain with the Biobricks I732011 (mRFP Tripart LVA-) don't to growth. | + | <br>We can only do the construction with mRFP Tripart (LVA+) because the stable strain with the Biobricks I732011 (mRFP Tripart LVA-) don't to growth. |
==Digestion== | ==Digestion== | ||
| Line 34: | Line 34: | ||
==='''Gel Verification'''=== | ==='''Gel Verification'''=== | ||
[[Team:Paris/Notebook/Protocols#Migration after extraction |Protocol]] | [[Team:Paris/Notebook/Protocols#Migration after extraction |Protocol]] | ||
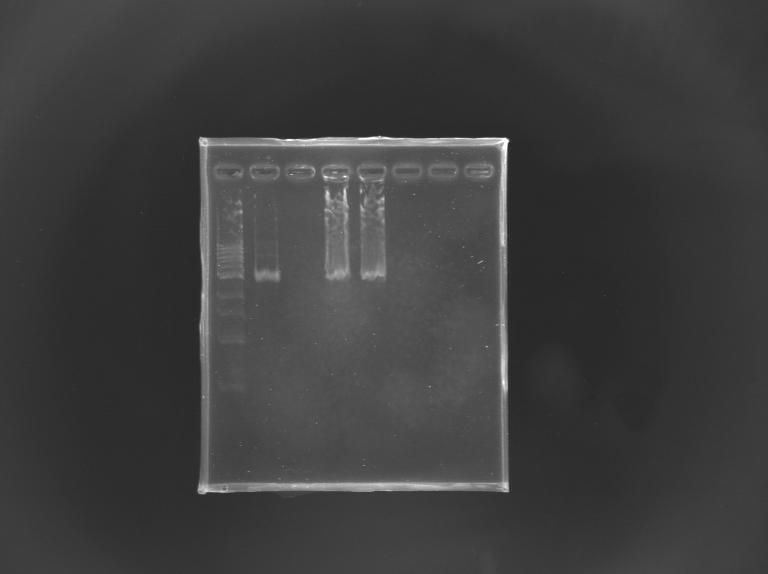
| - | [[Image: | + | [[Image:KR000246.JPG |thumb |Gel Verification of D187 digestion]] |
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| Line 54: | Line 54: | ||
|'''Expected size (pb)''' | |'''Expected size (pb)''' | ||
| | | | ||
| + | |2 955 | ||
| | | | ||
| - | | | + | |2 940 |
| - | + | ||
| colspan="2"| | | colspan="2"| | ||
|- | |- | ||
|'''Measured size (pb)''' | |'''Measured size (pb)''' | ||
| | | | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|2 900 |
| | | | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|2 900 |
|colspan="2"| | |colspan="2"| | ||
|} | |} | ||
| + | |||
=Cloning of EnvZ* in pSB1A2= | =Cloning of EnvZ* in pSB1A2= | ||
| Line 115: | Line 116: | ||
==Electrophoresis== | ==Electrophoresis== | ||
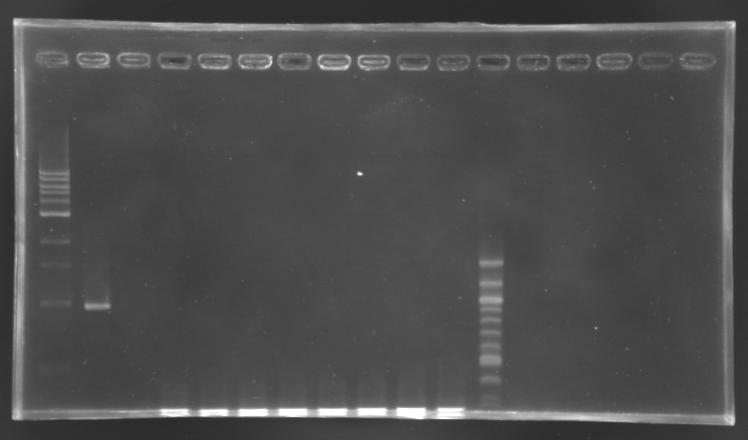
| - | [[Image:Screening EnvZ.JPG|thumb|]] | + | [[Image:Screening EnvZ.JPG|thumb|Screening of the cloning of EnvZ* - clones 1 to 8]] |
{|border="1" style="text-align:center" | {|border="1" style="text-align:center" | ||
| Line 134: | Line 135: | ||
|'''sample''' | |'''sample''' | ||
|1 kb DNA ladder | |1 kb DNA ladder | ||
| - | |positive control | + | |positive<br>control |
| - | |negative control | + | |negative<br>control |
| - | | | + | |L165.1 |
| + | |L165.2 | ||
| + | |L165.3 | ||
| + | |L165.4 | ||
| + | |L165.5 | ||
| + | |L165.6 | ||
| + | |L165.7 | ||
| + | |L165.8 | ||
|100 bp DNA ladder | |100 bp DNA ladder | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|- | |- | ||
|'''expected size''' | |'''expected size''' | ||
| Line 160: | Line 154: | ||
| | | | ||
|- | |- | ||
| - | |'''measured size''' | + | |style="background: #ff6d73" |'''measured size''' |
| | | | ||
| | | | ||
| | | | ||
| - | |colspan="8"| | + | |colspan="8"|0,3 kb |
| | | | ||
|- | |- | ||
| Line 171: | Line 165: | ||
No correct clone | No correct clone | ||
<br>The 8 other clones were also screened. | <br>The 8 other clones were also screened. | ||
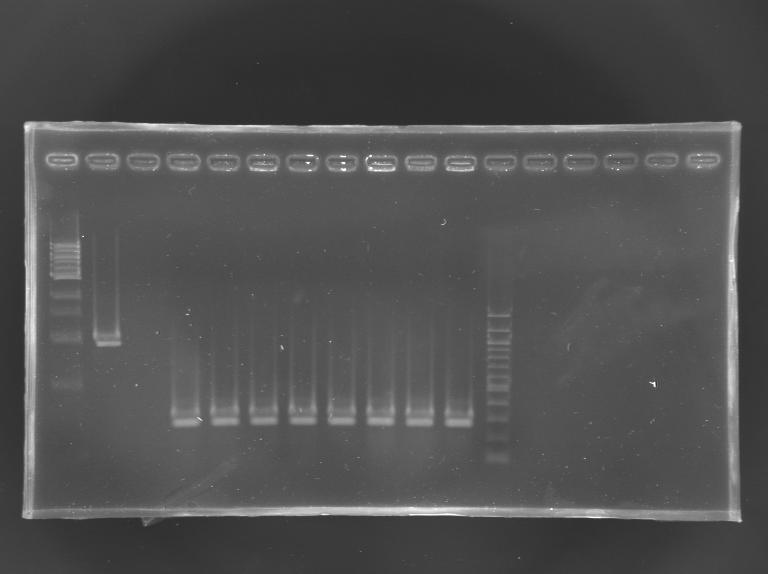
| - | [[Image:KR000256.JPG|thumb|]] | + | [[Image:KR000256.JPG|thumb|Screening of the cloning of EnvZ* - clones 9 to 16]] |
'''PCR''' | '''PCR''' | ||
elongation time: 2 min 30 | elongation time: 2 min 30 | ||
| Line 193: | Line 187: | ||
|'''sample''' | |'''sample''' | ||
|1 kb DNA ladder | |1 kb DNA ladder | ||
| - | |positive control | + | |positive<br>control |
| - | |negative control | + | |negative<br>control |
| - | | | + | |L165.9 |
| + | |L165.10 | ||
| + | |L165.11 | ||
| + | |L165.12 | ||
| + | |L165.13 | ||
| + | |L165.14 | ||
| + | |L165.15 | ||
| + | |L165.16 | ||
|100 bp DNA ladder | |100 bp DNA ladder | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|- | |- | ||
|'''expected size''' | |'''expected size''' | ||
| Line 219: | Line 206: | ||
| | | | ||
|- | |- | ||
| - | |'''measured size''' | + | |style="background: #ff6d73" |'''measured size''' |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| | | | ||
| | | | ||
| | | | ||
| + | |colspan="8"|0,3 kb | ||
| | | | ||
|- | |- | ||
|} | |} | ||
<br> | <br> | ||
| + | '''Results''': All the clones analysed were not correct. | ||
| + | |||
=Cloning of OmpR*= | =Cloning of OmpR*= | ||
| Line 296: | Line 278: | ||
*10 µL of template DNA or 10 µL of EB buffer for th blank | *10 µL of template DNA or 10 µL of EB buffer for th blank | ||
*50 µL of pure water | *50 µL of pure water | ||
| - | |||
{|border="1" style="text-align:center" | {|border="1" style="text-align:center" | ||
| Line 318: | Line 299: | ||
* 1 µL T4 DNA ligase at 400 000 U/mL concentration | * 1 µL T4 DNA ligase at 400 000 U/mL concentration | ||
* O/N at 16°C | * O/N at 16°C | ||
| + | |||
=Checking mutagenesis FliA= | =Checking mutagenesis FliA= | ||
| Line 323: | Line 305: | ||
For this, i digested mutated FliA and non-mutated FliA with EcoRI and PstI and put in migration the digestion products running on gels.<br> | For this, i digested mutated FliA and non-mutated FliA with EcoRI and PstI and put in migration the digestion products running on gels.<br> | ||
| - | Results : No digestion for the mutated sequence --> successful mission ! | + | Results : No digestion for the mutated sequence --> successful mission ! |
Latest revision as of 19:16, 9 September 2008
Construction of pFlhB - mRFP Tripart (LVA+)Aim : Construction of "pFlhB-RBS-mRFP-dbl ter" (pFlhB-I732078) DigestionMeasurement of the concentration of D187 purifiedProtocol (it's same that for Miniprep) => the experiments of Gel Extraction have failed, so we need to repeat the step of digestion. Digestion
Gel Verification
Cloning of EnvZ* in pSB1A2Transformation results
PCR screening
Electrophoresis
PCR
elongation time: 2 min 30
Results: All the clones analysed were not correct.
Cloning of OmpR*DigestionDetermination of the concentration of DNAWe used the biophotometer
Name of the digestions
Protocol of digestion
Cleaning of the digestion productsLigationDetermination of the concentration of DNAWe used the biophotometer
Protocol of ligation L171
Checking mutagenesis FliAFor this, i digested mutated FliA and non-mutated FliA with EcoRI and PstI and put in migration the digestion products running on gels. Results : No digestion for the mutated sequence --> successful mission ! |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"