Team:Paris/August 8
From 2008.igem.org
(Difference between revisions)
(→PCR Screening of Ligation Transformants) |
|||
| Line 83: | Line 83: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Transformation results== | ==Transformation results== | ||
| Line 255: | Line 145: | ||
|align="center"|'''fluorescence''' | |align="center"|'''fluorescence''' | ||
|- | |- | ||
| - | |rowspan="8"align="center"|L128 | + | | rowspan="8" align="center"|L128 |
| - | |rowspan="8"align="center"|pFlgA | + | | rowspan="8" align="center"|pFlgA |
|align="center"|1 | |align="center"|1 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|2 | |align="center"|2 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|3 | |align="center"|3 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|4 | |align="center"|4 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|5 | |align="center"|5 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|6 | |align="center"|6 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|7 | |align="center"|7 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|8 | |align="center"|8 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |rowspan="8"align="center"|L129 | + | | rowspan="8" align="center"|L129 |
| - | |rowspan="8"align="center"|pFlgB | + | | rowspan="8" align="center"|pFlgB |
|align="center"|1 | |align="center"|1 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|2 | |align="center"|2 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|3 | |align="center"|3 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|4 | |align="center"|4 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|5 | |align="center"|5 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|6 | |align="center"|6 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|7 | |align="center"|7 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|8 | |align="center"|8 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |rowspan="8"align="center"|L130 | + | | rowspan="8" align="center"|L130 |
| - | |rowspan="8"align="center"|pFlhB | + | | rowspan="8" align="center"|pFlhB |
|align="center"|1 | |align="center"|1 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|2 | |align="center"|2 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|3 | |align="center"|3 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|4 | |align="center"|4 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|5 | |align="center"|5 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|6 | |align="center"|6 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|7 | |align="center"|7 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|8 | |align="center"|8 | ||
|align="center"|no | |align="center"|no | ||
|- | |- | ||
| - | |rowspan="8"align="center"|L131 | + | | rowspan="8" align="center"|L131 |
| - | |rowspan="8"align="center"|pFlgB | + | | rowspan="8" align="center"|pFlgB |
|align="center"|1 | |align="center"|1 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|2 | |align="center"|2 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|3 | |align="center"|3 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|4 | |align="center"|4 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|5 | |align="center"|5 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|6 | |align="center"|6 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|7 | |align="center"|7 | ||
|align="center"|red | |align="center"|red | ||
|- | |- | ||
| - | |||
| - | |||
|align="center"|8 | |align="center"|8 | ||
|align="center"|no | |align="center"|no | ||
| Line 459: | Line 293: | ||
| - | === | + | * Program PCR "Screening": Annealing 55°C - Time élongation 1'30" - Number cycle : 29 |
| + | |||
| + | |||
| + | |||
| + | === Electrophoresis Purification of PCR=== | ||
| + | |||
| + | ''When the PCR cycles were finished,'' | ||
| + | |||
| + | '''conditions :''' | ||
| + | |||
| + | * 10µl of ladder 1 kb (unlike 100 pb) | ||
| + | * 2 x 30µl of PCR products added with 10µl of loading Dye 6x | ||
| + | * migration ~30min at 100W on a '''1,5% agarose gel'''. | ||
| + | |||
| + | |||
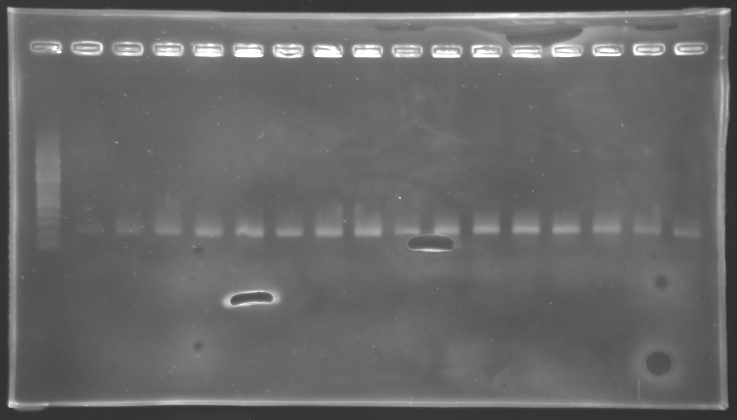
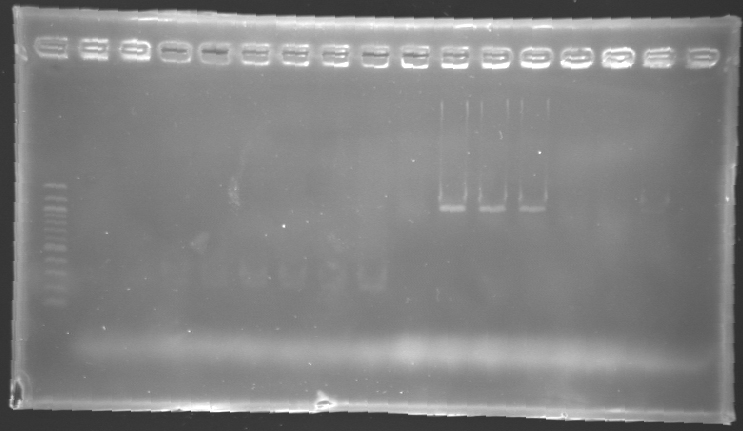
| + | '''Results of electrophoresis''' | ||
| - | + | <br>gel 1 [[Image:KR000143.jpg | gel 1|200px]] | |
| - | + | gel 2 [[Image:KR000145.jpg|gel 2|200px]] | |
| - | + | ||
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | {|- | + | {|- border="1" |
| - | |'''Name''' | + | |align="center"|'''Name''' |
| - | |'''Promotor''' | + | |align="center"|'''Promotor''' |
|align="center"|'''Gel''' | |align="center"|'''Gel''' | ||
|align="center"|'''Band''' | |align="center"|'''Band''' | ||
| Line 478: | Line 321: | ||
|align="center"|'''Measured size''' | |align="center"|'''Measured size''' | ||
|- | |- | ||
| - | |align="center"|PCR_124 | + | |align="center"|PCR_124' |
|align="center"|pFlgA | |align="center"|pFlgA | ||
|align="center"|1 | |align="center"|1 | ||
| - | |align="center"|2 | + | |align="center"|2 to 9 |
|style="background: #cbff7B"|<center>261 pb</center> | |style="background: #cbff7B"|<center>261 pb</center> | ||
| - | |align="center"| | + | |align="center"|300 pb |
|- | |- | ||
| - | |align="center"|PCR_125 | + | |align="center"|PCR_125' |
|align="center"|pFlgB | |align="center"|pFlgB | ||
|align="center"|1 | |align="center"|1 | ||
| - | |align="center"| | + | |align="center"|10 to 17 |
|style="background: #cbff7B"|<center>261 pb</center> | |style="background: #cbff7B"|<center>261 pb</center> | ||
| - | |align="center"| | + | |align="center"|300 pb |
|- | |- | ||
| - | |align="center"|PCR_126 | + | |align="center"|PCR_126' |
|align="center"|pFlhB | |align="center"|pFlhB | ||
| - | |align="center"| | + | |align="center"|2 |
| - | |align="center"| | + | |align="center"|2 to 9 |
|style="background: #cbff7B"|<center>260 pb</center> | |style="background: #cbff7B"|<center>260 pb</center> | ||
| - | |align="center"| | + | |align="center"|300 pb |
|- | |- | ||
| - | |align="center"|PCR_127 | + | |align="center"|PCR_127' |
|align="center"|pFlhDC | |align="center"|pFlhDC | ||
|align="center"|2 | |align="center"|2 | ||
| - | |align="center"| | + | |align="center"|10 to 17 |
|style="background: #ff6d73"|<center>446 pb</center> | |style="background: #ff6d73"|<center>446 pb</center> | ||
| - | |align="center"| | + | |align="center"|1,000 pb |
|} | |} | ||
| - | + | ||
| - | + | ||
| - | + | ==> '''Conclusion:''' | |
| - | * | + | *PCR of pFlgA, pFlgB and pFlhB have succeed, but we always have a problem with pFlhDC probably because of the oligos wich are not specific. |
Revision as of 10:29, 11 August 2008
|
Minipreps : Plasmid extractionExtraction of pSB3K3 et E0240 in pSB1A2 plasmid from overnight bacteria culture using the QIAspin Miniprep Kit (QIAGEN) by QIACube.
Amplification of Genes of interest (OmpR, EnvZ, FlhDC)We performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR Protocol
Transformation results
PCR Screening of Ligation TransformantsUse of 8 clones of Ligation transformants for screening PCR
Protocol of screening PCR
Conditions of electrophoresis
Electrophoresis Purification of PCRWhen the PCR cycles were finished, conditions :
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"