Team:Paris/August 21
From 2008.igem.org
AnaJimenez (Talk | contribs) (→Ligation) |
AnaJimenez (Talk | contribs) (→Ligation) |
||
| Line 487: | Line 487: | ||
| - | = | + | =Promoter characterization plasmids= |
| + | ==Ligations from digestions from 20th== | ||
| - | [[Team:Paris/Notebook/Protocols# | + | Top 10 cells were used |
| + | |||
| + | |||
| + | [[Team:Paris/Notebook/Protocols#Transformation |Protocol]] | ||
| Line 497: | Line 501: | ||
|'''Vector digestion''' | |'''Vector digestion''' | ||
|'''Vector description''' | |'''Vector description''' | ||
| + | |'''Vector conc. ug/mL''' | ||
|'''Vector volume''' | |'''Vector volume''' | ||
|'''Insert digestion''' | |'''Insert digestion''' | ||
|'''Insert description''' | |'''Insert description''' | ||
| + | |'''Insert conc. ug/mL''' | ||
|'''Insert volume''' | |'''Insert volume''' | ||
|'''Product description''' | |'''Product description''' | ||
| Line 507: | Line 513: | ||
|D164 | |D164 | ||
|J23101 promoter | |J23101 promoter | ||
| - | | | + | |3 |
| + | |16 | ||
|D163 | |D163 | ||
|gfp generator | |gfp generator | ||
| - | | | + | |14 |
| + | |8 | ||
|J23101 promoter-gfp generator | |J23101 promoter-gfp generator | ||
|Amp | |Amp | ||
| Line 517: | Line 525: | ||
|D161 | |D161 | ||
|pTet promoter | |pTet promoter | ||
| - | |1 | + | |39 |
| + | |1.25 | ||
|D163 | |D163 | ||
|gfp generator | |gfp generator | ||
| - | | | + | |14 |
| + | |5 | ||
|pTet promoter-gfp generator | |pTet promoter-gfp generator | ||
|Kana | |Kana | ||
|- | |- | ||
| - | | | + | |Control L156 |
|D161 | |D161 | ||
| | | | ||
| - | |1 | + | | |
| + | |1.25 | ||
| + | |none | ||
| | | | ||
| | | | ||
| Line 537: | Line 549: | ||
|D125.2 | |D125.2 | ||
|B0015 | |B0015 | ||
| + | |15 | ||
|3 | |3 | ||
|D162 | |D162 | ||
| - | |||
|tetR | |tetR | ||
| + | |11 | ||
| + | |4 | ||
|tetR-B0015 | |tetR-B0015 | ||
| + | |Kana | ||
| + | |- | ||
| + | |Control L157 | ||
| + | |D125.2 | ||
| + | | | ||
| + | | | ||
| + | |3 | ||
| + | |none | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |Vector autoligation control | ||
| + | |Kana | ||
| + | |- | ||
| + | |L166 | ||
| + | |D185 | ||
| + | |RBS B0032 | ||
| + | |21 | ||
| + | |2 | ||
| + | |D182 | ||
| + | |tetR | ||
| + | |10 | ||
| + | |6.5 | ||
| + | |RBS B0032 - tetR | ||
|Amp | |Amp | ||
|- | |- | ||
| + | |Control L166 | ||
| + | |D185 | ||
| | | | ||
| - | |||
| | | | ||
| + | |2 | ||
| + | |none | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |Vector autoligation control | ||
| + | |Amp | ||
| + | |- | ||
| + | |L167 | ||
| + | |D181 | ||
| + | |pTet | ||
| + | |1 | ||
| + | |14 | ||
| + | |D184 | ||
| + | |gfp generator | ||
| + | |14 | ||
|3 | |3 | ||
| + | |gfp generator - pTet | ||
| + | |Amp | ||
| + | |- | ||
| + | |Control L167 | ||
| + | |D181 | ||
| + | |pTet | ||
| + | | | ||
| + | |14 | ||
| + | |none | ||
| | | | ||
| | | | ||
Revision as of 16:55, 26 August 2008
Cloning of FlhB promoterProtocol
Resuspension of 1 colony E.coli K12 strain MG 1655 in 100µl of water.
1µl of dNTP
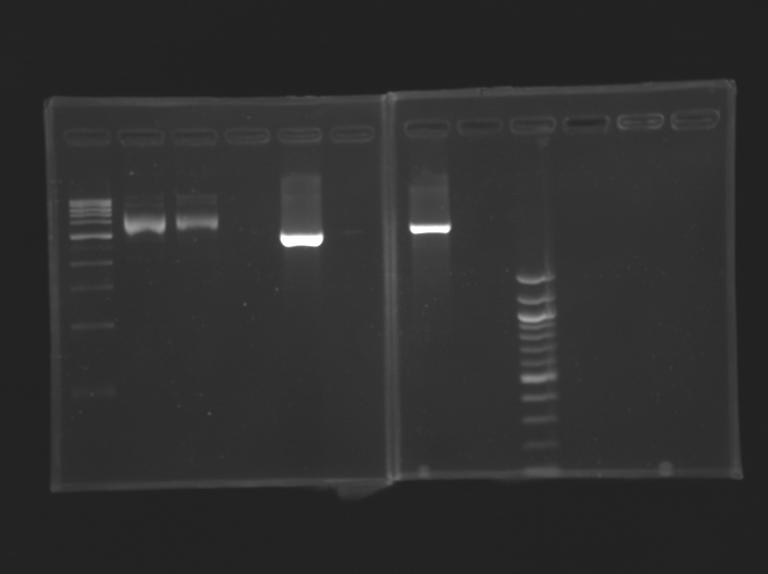
Result[[Image:KR00019b.jpg| thumb| Vérification of pFlhB]
Construction for FIFOAim : Construction of pFlgA - YFP tripart (+/- LVA) "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432) DigestionDigestion
Gel Extraction
Measurement of the concentration of D166 & D167 purifiedProtocol (it's same that for Miniprep)
Ligation
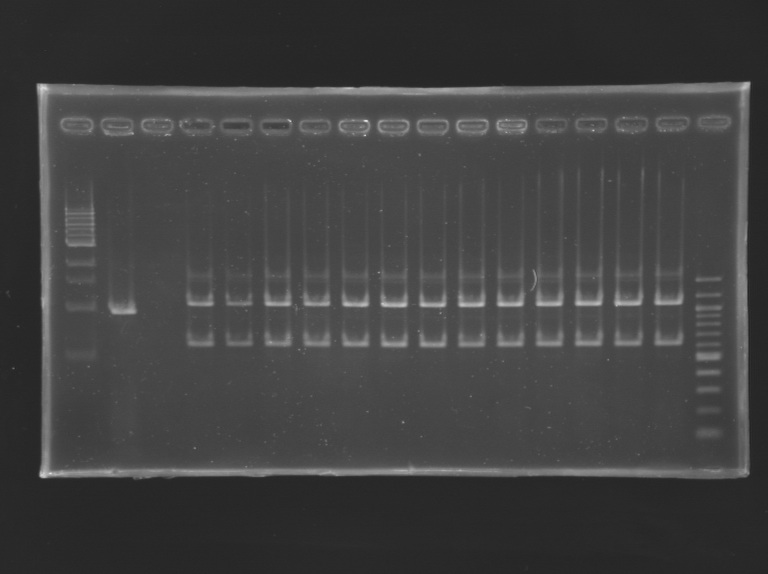
Screening of the cloning of E0240 and FlhDC+promotorWe obtained single colonies with the dilution 100.
PCR screeningreaction mixture (25 µL)
PCR screening programm
Electrophoresis
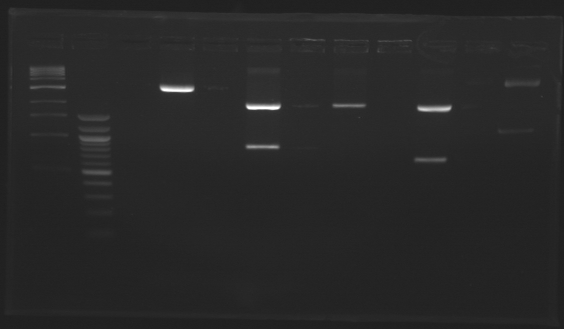
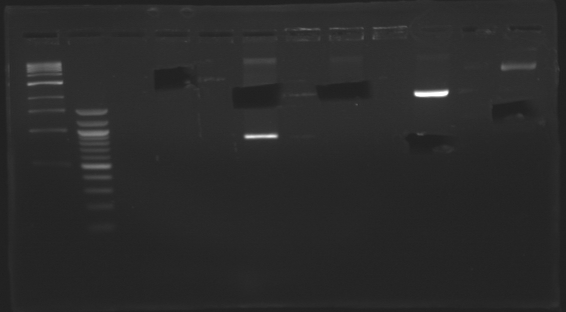
Construction for synchronizationLigations
Promoter characterization plasmidsLigationOur ligations from yesterday didn't work. The positive control for transformation worked. DigestionWe had a problem with a gel extraction so we have to make again the digestions from yesterday
Promoter characterization plasmidsLigations from digestions from 20thTop 10 cells were used
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"