From 2008.igem.org
(Difference between revisions)
|
|
| Line 191: |
Line 191: |
| | |Amp | | |Amp |
| | |} | | |} |
| - |
| |
| | | | |
| | | | |
Revision as of 13:22, 30 August 2008
|
← Yesterday ↓ Calendar ↑Tomorrow →
Construction of OmpR*+RBS and EnvZ*+RBS: Ligations
Cleaning of the DNA after the digestion
We used the QIAcube to wash the DNA, following the standard protocol.
Measure of DNA concentration of the digestion products
We used the biophotometer.
Settings:
- 10 µL of template DNA in 50 µL of pure water
- Blank : 10 µL of EB buffer in 50 µL of water.
| Digestion name
| What's in ?
| DNA concentration (ng/µL)
|
| D 158
| OmpR*
| 12
|
| D 159
| EnvZ*
| 7
|
| D 102
| B0034
| 16
|
List of ligations
- We ligated the DNA following the standard protocol.
- T5 is the autoligation control for L148 and L149.
| Ligation name
| Insert name
| Vinsert µL
| Vector name
| VVector µL
|
| L 148 (Amp)
| D 158 (OmpR*)
| 4
| D 102 (B0034)
| 3
|
| L 149 (Amp)
| D 159 (EnvZ)
| 7
| D 102 (B0034)
| 3
|
Creation of a registry of pFliL, pFlhDC, and FlhDC
Analysis of the transformation we did yesterday
L143, L144, T1 and T2 showed no colonies. The positive control with pUC19 worked well.
We suppose that the ligation did not work. We will do it again today.
Cleaning of the DNA after the digestion
We used the QIAcube to wash the DNA, following the standard protocol.
Measure of DNA concentration of the digestion products
We used the biophotometer.
Settings:
- 10 µL of template DNA in 50 µL of pure water
- Blank : 10 µL of EB buffer in 50 µL of water.
| Digestion name
| What's in ?
| DNA concentration (ng/µL)
|
| D 149
| pfliL
| 2
|
| D 150
| pfliL
| 3
|
| D 151
| pSB3K3
| 12
|
| D 152
| pSB3K3
| 20
|
| D 153
| gene flhDC
| 7
|
| D 154
| gene flhDC
| 12
|
| D 155
| pflhDC
| 16
|
| D 136
| j61002
| 12
|
| D 145
| pSB1A2
| 21
|
List of ligations
- We ligated the DNA following the standard protocol.
- T1, T2, T3 and T4 are the autoligation controls for L 143, L144, L145 and L147
| Ligation name
| Insert name
| Vinsert µL
| Vector name
| VVector µL
|
| L 143 (Kan)
| D 149 (pfliL)
| 8
| D 137 (pSB3K3)
| 4
|
| L 144 (Kan)
| D 150 (pfliL)
| 5
| D 152 (pSB3K3)
| 2.5
|
| L 145 (Amp)
| D 153 (g flhDC)
| 10
| D 145 (pSB1A2)
| 2.5
|
| L 146 (Amp)
| D 154 (g flhDC)
| 5
| D 145 (pSB1A2)
| 2.5
|
| L 147 (Amp)
| D 155 (p flhDC)
| 1
| D 136 (J61002)
| 4
|
Construction of pLas-TetR-GFP tripart & rbs-LasR-dble ter
Transformation
Transformation protocol
| Name
| Description
| Antibio
|
| Ligations
|
| L150
| D105(BV) - D146(BI)
pLas - strongest rbs-TetR-GFP tripart
| Amp
|
| L151
| D147(FI) - D125 (FV)
strongest rbs-LasR activator with LVA - Double terminator
| Kan
|
| Controls
|
| C1
| D105
| Amp
|
| C2
| D125
| Kan
|
| Positive Control
| pUC19
| Amp
|
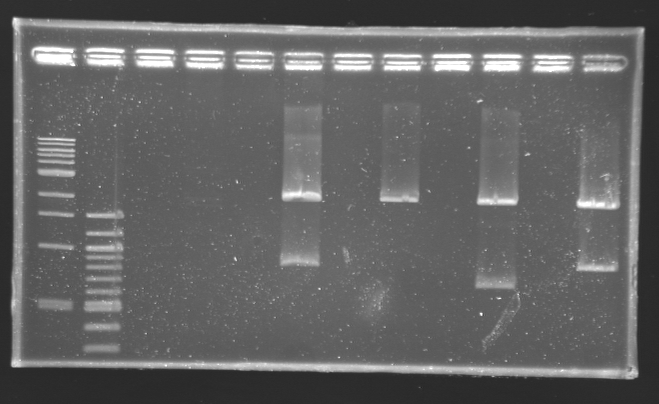
Digestion check from yesterday
Protocol Digestion
| Digestion name
| Plasmid
| Description
| Miniprep used
| Comments
|
|
| MP3
| B0015 (double terminator B0010-B0012) - FV
| 4
| No bands
|
| D164
| MP101
| promoter J23101 - BV
| 1
| Good expected sized bands
|
| D161
| MP104
| PTet (Tet promoter) - FI
| 1
| Good expected sized band
|
| D162
| MP114
| TetR - BI
| 1
| Good expected sized bands
|
| D163
| MP143
| gfp generator - BV
| 2
| Good expected sized bands
|
|
 "
"