Team:Paris/August 12
From 2008.igem.org
(Difference between revisions)
(→Minipreps: Plasmid extraction) |
(→Protocol) |
||
| Line 39: | Line 39: | ||
|align="center"|'''Volume of DNA''' | |align="center"|'''Volume of DNA''' | ||
|- | |- | ||
| - | |align="center"|D | + | |align="center"|D 140 |
| - | |align="center"| | + | |align="center"|PCR 130 - E0240 RBS + |
| - | |align="center"| PstI | + | |align="center"|PstI |
| - | |align="center"| | + | |align="center"|5 µL |
|- | |- | ||
| - | |align="center"|D | + | |align="center"|D 141 |
| - | |align="center"| | + | |align="center"|PCR 131 - flhD rbs - |
|align="center"|EcoRI-SpeI | |align="center"|EcoRI-SpeI | ||
| - | |align="center"| | + | |align="center"|5 µL |
| + | |- | ||
| + | |align="center"|D 142 | ||
| + | |align="center"|PCR 132 - flhC rbs - | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|5 µL | ||
| + | |- | ||
| + | |align="center"|D 143 | ||
| + | |align="center"|PCR 133 - flhDC + prom | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|5 µL | ||
| + | |- | ||
| + | |align="center"|D 144 | ||
| + | |align="center"|MP 142 - pSB3K3 | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|5 µL | ||
| + | |- | ||
| + | |align="center"|D 145 | ||
| + | |align="center"|MP122 - pSB1A2 | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|5 µL | ||
|} | |} | ||
| Line 57: | Line 77: | ||
* Incubate during about 3h at 37°C, then 20 minutes at 65°C (to inactivate the enzymes). | * Incubate during about 3h at 37°C, then 20 minutes at 65°C (to inactivate the enzymes). | ||
| + | * Exclusively for D 144 : 2 µL of Antarctic Phosphatase was added, 30 min at 37°C then 5 min at 65°C. | ||
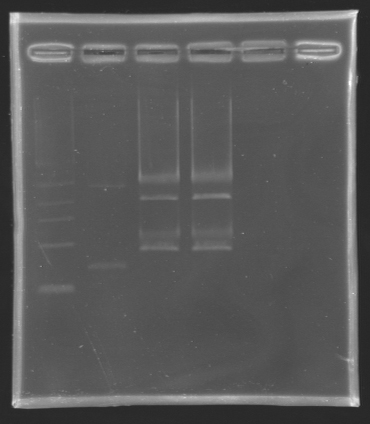
====Results of the digestion==== | ====Results of the digestion==== | ||
Revision as of 14:29, 14 August 2008
|
Digestion of the PCR we did yesterdayYesterday we amplified flhD, flhC, flhDC with its promoter and RBS+ Before the digestion, we have to determine the DNA concentration of the templates. Measurement of DNA concentrationWe used the biophotometer. We put 5 µL of template DNA in 95 µL of water. The Blank was made with 5 µL of EB Buffer (the one that was used for the elution of the DNA) in 95 µL of water.
DigestionProtocol
Results of the digestionElectrophoresis settings
Gel extraction and DNA purificationTo extract the biobrick E0240 from the gel, we used the standard protocol number 8. The ligation will be done tomorrow. Minipreps: Plasmid extraction
Glycerol Stocks
Results of the transformations we did yesterdayYour results please Cyprien!!!??? Concentration of the MiniPreps
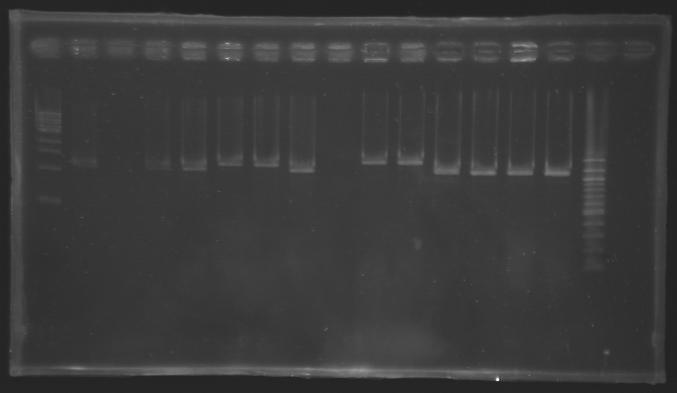
New PCR screening with the right primersTransformants with pFlgA, pFlgB and pFlhB cloned into J61002 are analysed by PCR but this time with the right primers: VF2 (O18) and VR (O19).
Electrophoresis
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"