| Ligation name
| Description
| Antibio
| Nb colonies
| Comments
|
| L100
| rbs TetR - ECFP
D110 (BV) - D130 (BI)
| Amp
| 0
| After another night nothing was osberved
-> To do again
|
| L101
| rbs TetR - GFP tripart
D110 (BV) - D131 (BI)
| Amp
| 1
| After another night nothing was observed
-> To do again
|
| L102
| Strong rbs - YFP
D129 (BV) - D118 (BI)
| Amp
| +/- 100
|
|
| L103
| Strong rbs - GFP
D129 (BV) - D122 (BI)
| Amp
| a lot
|
|
| L104
| Strong rbs - lasR activator
D129 (BV) - D114 (BI)
| Amp
| a lot
|
|
| L105
| Strong promoter - ECFP
D123 (BV) - D130 (BI)
| Amp
| a lot
|
|
| L106
| Strong promoter - gfp Tripart
D123 (BV) - D131 (BI)
| Amp
| a lot
|
|
| L107
| Strongest promoter - ECFP
D103 (BV) - D131 (BI)
| Amp
| a lot
|
|
L108 n°2
(the right one)
| Strong promoter - gfp Tripart
D103 (BV) - D130 (BI)
| Amp
| +/- 100
|
|
| L109 n°1
| Strong promoter - ecfp
D124 (BV) - D130 (BI)
| Amp
| +/- 30
| To do again
|
| L109 n°2
| Strong promoter - ecfp
D124 (BV) - D130 (BI)
| Amp
| a lot
| To do again
|
| L110
| Medium promoter - gfp Tripart
D124 (BV) - D131 (BI)
| Amp
| a lot
|
|
| L111
| Weak promoter - ECFP
D104 (BV) - D130 (BI)
| Amp
| a lot
|
|
| L112
| Weak promoter - gfp
D104 (BV) - D131 (BI)
| Amp
| a lot
|
|
| L113
| AracpBAD - ecfp
D126 (BV) - D130 (BI)
| Kan
| 1
| After another night nothing was observed
-> To do again
|
| L114
| AracpBAD - gfp tripart
D126 (BV) - D131 (BI)
| Kan
| 2
| After another night nothing was observed
-> To do again
|
| L115
| pLas - ECFP
D105 (BV) - D130 (BI)
| Amp
| a lot
|
|
| L116
| pLas - gfp Tripart
D105 (BV) - D131 (BI)
| Amp
| a lot
|
|
| L117
| yfp - Double Terminator
D117 (FI) - D125 (FV)
| Amp
| +/- 30
|
|
| L118
| rfp - Double Terminator
D121 (FI) - D125 (FV)
| Amp
| +/- 30
|
|
| L119
| lasR activator + LVA - Double Terminator
D113 (FI) - D125 (FV)
| Amp
| +/- 30
|
|
| L120
| tetR repressible - ECFP
D106 (BV) - D130 (BI)
| Amp
| a lot
|
|
| L121
| Strong promoter - gfp tripart
D106 (BV) - D130 (BI)
| Amp
| a lot
|
|
| L122
| RBS-lasI - ecfp
D107 (BV) - D130 (BI)
| Amp
| 0
| After another night nothing was observed
-> To do again
|
| L123
| RBS lasI - ecfp
D107 (BV) - D131 (BI)
| Amp
| 1
| After another night nothing was observed
-> To do again
|
| L124
| Strongest RBS - rfp
D102 (BV) - D122 (BI)
| Amp
| a lot
|
|
| L125
| Strongest RBS - yfp
D102 (BV) - D118 (BI)
| Amp
| a lot
|
|
| L126
| Strongest RBS (1)- LacR activator (+LVA)
D102 (BV) - D118 (BI)
| Amp
| no dish found!!!
| To do again
|
| L127
| gfp (1)- Double terminator
D119 (FI) - D125 (FV)
| Amp
| no dish found!!!
| To do again
|
| Controls
|
|
|
|
|
| C1
|
D110
| Amp
| 0
| -
|
| C2
|
D129
| Amp
| 5
| -
|
| C3
|
D123
| Amp
| +/- 100
| -
|
| C4
|
D103
| Amp
| +/- 20
| -
|
| C5
|
D124
| Amp
| +/- 100
| -
|
| C6
|
D104
| Amp
| +/- 100
| -
|
| C7
|
D126
| Kana
| 0
| -
|
| C8
|
D105
| Amp
| +/- 20
| -
|
| C9
|
D125
| Amp
| 0
| -
|
| C10
|
D106
| Amp
| 0
| -
|
| C11
|
D107
| Amp
| 0
| -
|
| C12
|
D102
| Amp
| +/- 10
| -
|
| Positive control
|
puc19
| Amp
| 155 (transformation
efficiency:1.5*10^7/ug)
| -
|
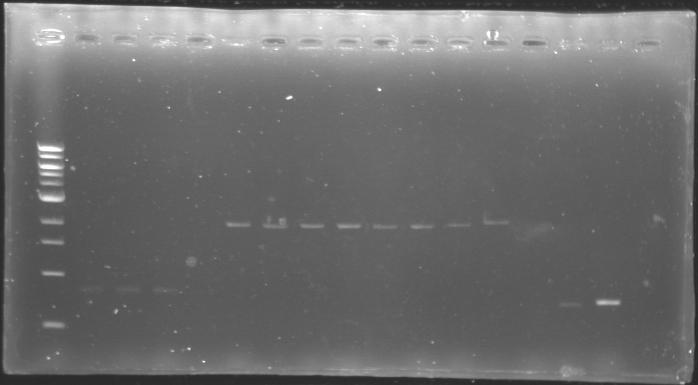
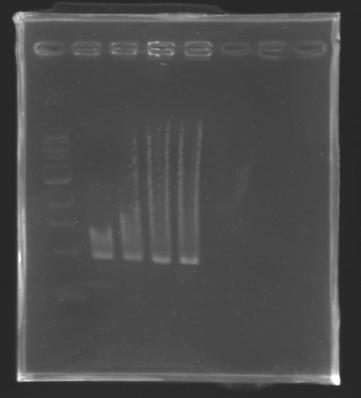
The digested DNA of yesterday was analysed one more time by electrophoresis on a 0.8% agarose gel (about 30 minutes at 100 W). The ladder used was the 1 kb DNA ladder (New England Biolabs). 5 µL of each sample with 1 µL of loading dye were loaded.
| Name
| BioBrick
| Tube N°
| Enz 1
| Enz 2
| Obs
| Exp Size
| Mea Size
| Conc (ng/µl)
| Band
|
| D101
| B0034
| 1
| EcoRI
| XbaI
| FV
| 2076 bp
| 2000 bp
| 6
| 10
|
| D102
| B0034
| 1
| SpeI
| PstI
| BV
| 2077 bp
| 2000 bp
| 8
| 11
|
| D105
| R0079
| 1
| SpeI
| PstI
| BV
| 2222 bp
| 2000 bp
| 6
| 13
|
| D106
| R0040
| 1
| SpeI
| PstI
| BV
| 2119 bp
| 2000 bp
| 6
| 12
|
| D108
| S03154
| 1
| XbaI
| PstI
| BI
| 707 bp
|
| 0 (failed)
| 14
|
| D111
| S03879
| 1
| XbaI
| PstI
| BI
| 725 bp
| 600 bp
| 2
| 15
|
| D115
| C0179
| 2
| EcoRI
| SpeI
| FI
| 746 bp
|
| 0 (failed)
| 4 & 5
|
| D116
| C0179
| 2
| XbaI
| PstI
| BI
| 745 bp
| 700 bp
| 2
| 2 & 3
|
| D117
| E0030
| 1
| EcoRI
| SpeI
| FI
| 746 bp
| 600 bp
| 10
| 16
|
| D128
| B0030
| 2
| EcoRI
| XbaI
| FV
| 2079 bp
| 2000 bp
| 8
| 6 & 7
|
| D129
| B0030
| 2
| SpeI
| PstI
| BV
| 2080 bp
| 2000 bp
| 8
| 8 & 9
|
Results :
Each of the samples was succesfully digested and purified except for the sample D108. It seems that the QIAprep columms (from the QIAGEN Minipreps kit) can be used instead of the QIAquick columms (for DNA Gel Extraction).
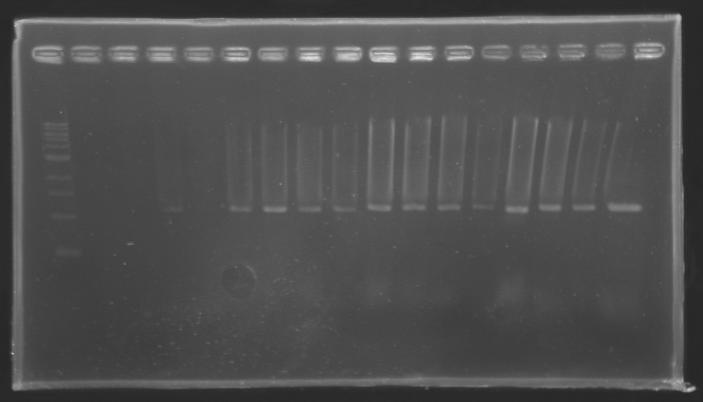
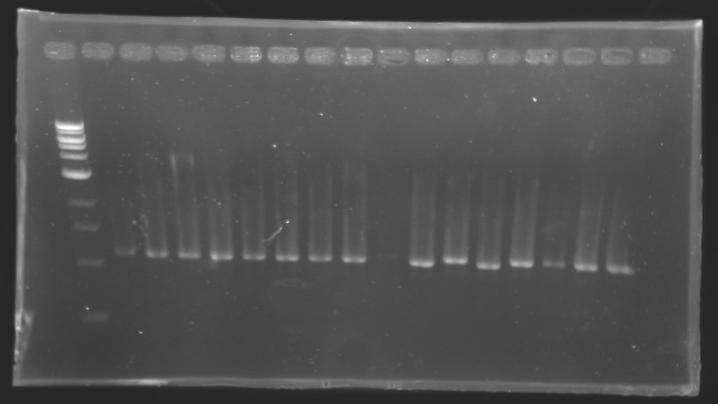
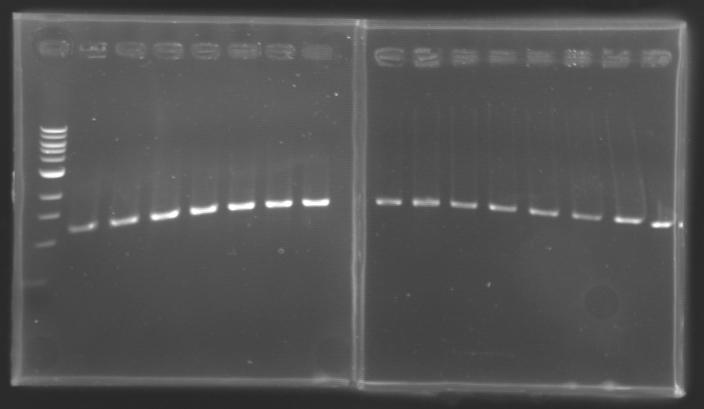
PCR Screening of Ligation Transformants
Use of 8 clones of Ligation transformants for screening PCR
Protocol of screening PCR
| Name
| Vol (µl)
| Concentration
|
| Quick Load
| 25µl
| 2X
|
| OligoF_VF2 (O18)
| 1µl
| 10µM
|
| OligoR_VR (O19)
| 1µl
| 10µM
|
| water
| 23µl
|
- 50µl of Mix PCR by tube/clone
- one toothpick of each clone's colony by tube
- Program : Annealing 55°C - Time élongation 1'30" - Number cycle : 29
Conditions of electrophoresis
- 10µl of ladder 1 kb
- 15µl of screening PCR (gel n°1, 2, 3(9-17), 4, 5, 6, 7, 8, 9, 10, 11)
- 10µl of screening PCR (gel n°3(1), 13, 14)
- migration ~30min at 100W on 0,8% gel
Results
| PCR1_’’’L102(1-8)’’’
| PCR2_’’’L103(1-8)’’’
| PCR3_’’’L104(1-8)’’’
| PCR4_’’’L105(1-8)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1045 pb
| 1000 pb
| 2-->9
| 1003 pb
| 1000 pb
| 10-->17
| 1078 pb
| 1000pb
| 2-->9
| 1239 pb
| 1200 pb
| 10-->17
|
|
|
|
| PCR5_’’’L106(1-8)’’’
| PCR6_’’’L107(1-8)’’’
| PCR7_’’’L108.1(1-8)’’’
| PCR8_’’’L108.2(1-8)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1200 pb
| 1200 pb
| 2-->9
| 1239 pb
| 1200 pb
| 10-->17
| 1200 pb
| 1200 pb
| 2-->9
| 1200 pb
| 1200 pb
| 10-->17
|
|
|
|
| PCR9_’’’L110(1-8)’’’
| PCR10_’’’L111(1-8)’’’
| PCR11_’’’L112(1-8)’’’
| PCR12_’’’L115(1-7)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1200 pb
| 1100 pb
| 2-->9
| 1239 pb
| 1100 pb
| 10-->17
| 1200 pb
| 1000 pb
| 2-->10
| 1239 pb
| 700 pb
| 11-->16
|
|
|
|
| PCR13_’’’L115(8)’’’
| PCR14_’’’L116(1-8)’’’
| PCR15_’’’L117(1-6)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1239 pb
| 1200 pb
| 2
| 1200 pb
| 1200 pb
| 3-->10
| 1046 pb
| 1000 pb
| 11-->16
|
|
|
| PCR16_’’’L117(7-8)’’’
| PCR17_’’’L118(1-8)’’’
| PCR18_’’’L119(1-5)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1046 pb
| 1000 pb
| 2-->3
| 1004 pb
| 1100 pb
| 4-->11
| 1079 pb
| 1200 pb
| 12-->16
|
|
|
| PCR19_’’’L119(6-8)’’’
| PCR20_’’’L120(1-8)’’’
| PCR21_’’’L121(1-4)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1079 pb
| 1100 pb
| 2-->4
| 1239 pb
| 1000 pb
| 5-->12
| 1200 pb
| 1200 pb
| 13-->16
|
|
|
| PCR22_’’’L121(5-8)’’’
| PCR23_’’’L122(1-8)’’’
| PCR24_’’’L125(1-4)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1200 pb
| 1000 pb
| 2-->5
| 1239 pb
| 1000 pb
| 6-->12
| 1045 pb
| 1000pb
| 13-->16
|
|
|
| PCR25_’’’L125(5-8)’’’
| PCR26_’’’L109.1(1-7)’’’
| PCR27_’’’L109.2(1-8)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1045 pb
| 1000 pb
| 2-->5
| 1239 pb
| 1100 pb
| 2-->9
| 1239 pb
| 1100 pb
| 10-->17
|
|
|
|
==> Conclusion : with the PCR, we have check that the transformant bacteria contain insert. (obtain amplification at the good size).
But we don't observe results for L102(3), L102(6), L103(4), L106(1), L106(2), L106(4), L111(1)
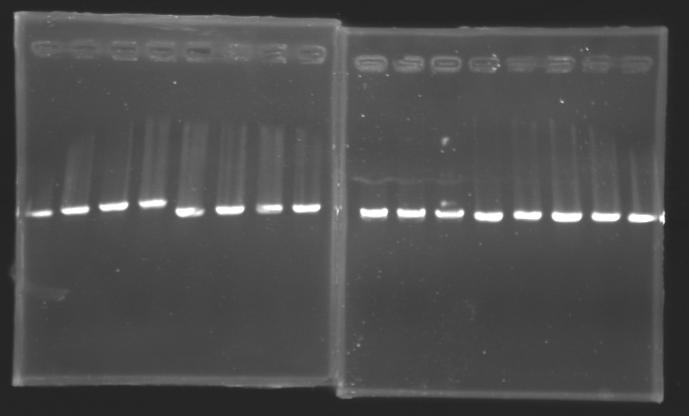
Migration of an another gel for this sample...
Results:
| PCR1_’’’L102(3;6))’’’
| PCR2_’’’L103(4)’’’
| PCR5_’’’L106(1; 2; 4)’’’
| PCR10_’’’L111(1)’’’
|
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
| Expected size
| Measured size
| Band
|
| 1045 pb
| 1000 pb
| 2-3
| 1003 pb
| -
| 4
| 1200 pb
| 1100 pb
| 5-6-7
| 1239 pb
| 1100 pb
| 8
|
 Gel 13 : to solve Mistakes |
==> Conclusion : We can observe a results for the samples : L102, L103, L106(4) and L111.
(but not for L106(1; 2))
|
 "
"