|
← Yesterday ↓ Calendar ↑Tomorrow →
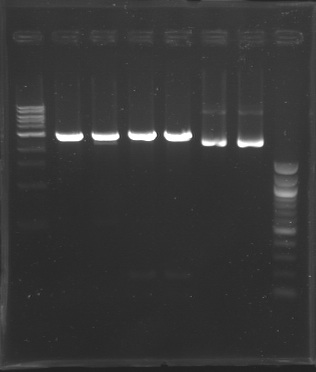
Screening of the cloning of E0240 and FlhDC+promotor
Spreading the clones in order to obtain single colonies
| Strain
| Resistance
| Ligation
| DNA cloned
| vector
| expected size of the fragment amplified by VF & VR
| mesured size
|
| S159.1
| kanamycine
| L139.1
| E0240 (GFP tripart)
| pSB3K3
| 1192 bp
| 1,5 kb
1,1 kb
0,6 kb
|
| S161.1
| ampicilline
| L142.7
| FlhDC+promotor
| pSB1A2
| 1403 bp
| 1,4
0,4 kb
0,3 kb
|
The plates obtained from the speading of yesterday can't be used because there are not single colonies.
We have to try again, but with a stronger dilution of the bacteria or with a smaller volume of spreading.
- Resuspension of some bacteria from the glycerol stock into 1 mL of LB+antibiotic
- Dilution 10 and dilution 100
- Spreading of 100 µL of each dilution on a LB plate containing the right antibiotic
- Overnight incubation (37°C)
Miniprep and stock glycerol
| Stock number
| Miniprep number
| Biobricks
| Description
|
| S163.1
| MP163.1
| B0032
| 
RBS
|
| S163.2
| MP163.2
|
| S164.1
| MP164.1
| E0422
|    
RBS+ ECFP+ LVA+ term
|
| S164.2
| MP164.2
|
| S165.1
| MP165.1
| E0430
|    
RBS+ YFP+ LVA- term
|
| S165.2
| MP165.2
|
| S166.1
| MP166.1
| E0432
|    
RBS+ YFP LVA+ term
|
| S166.2
| MP166.2
|
| S167.1
| MP167.1
| E0420
|    
RBS+ ECFP LVA- term
|
| S167.2
| MP167.2
|
| S168.1
| MP168.1
| I732078
|    
RBS+ mRFP LVA+ term
|
| S168.2
| MP168.2
|
Construction for FIFO
Aim : Construction of pFlgA - YFP tripart (+/- LVA) "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432)     
Digestion
Measurement of concentration of minipreps
standard protocol
| Miniprep
| Biobrick
| C° (µg/mL)
| ratio 260/280
|
| MP164.1
| E0422
| 95
| 1.69
|
| MP164.2
| E422
| 90
| 1.76
|
| MP165.1
| E0430
| 131
| 1.74
|
| MP165.2
| E0430
| nd
| nd
|
| MP166.1
| E0432
| 112
| 1.65
|
| MP166.2
| E0432
| 79
| 1.6
|
| MP167.1
| E0420
| 199
| 1.72
|
| MP167.2
| E0420
| 194
| 1.73
|
| MP168.1
| I732078
| 111
| 1.65
|
| MP168.2
| I732078
| 104
| 1.67
|
| MP122.1
| E0840
| 56
| 1.58
|
| MP122.2
| E0840
| 98
| 1.63
|
Digestion
Protocol Digestion
| Name
| Template DNA
| Description
| Vol MP (µl)
| Vol H2O (µl)
| Enzymes
|
| D166
| MP165.1
| RBS+ YFP LVA- term - FV
| 7.63
| 17
| EcoRI and XbaI
|
| D167
| MP166.1
| RBS+ YFP LVA+ term - FV
| 8.9
| 15.8
| EcoRI and XbaI
|
| D131
| MP122.2
| GFP tripart - I
| 10.2
| 14.5
| XbaI and PstI
|
Protocol
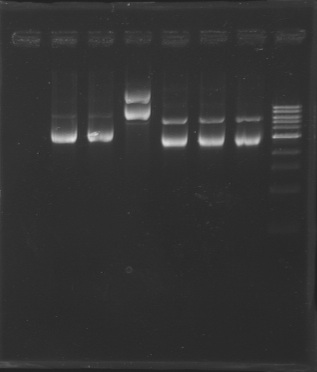
 Gel Extraction of D166-D167-D131
| Well
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
|
| Sample
| 1kb ladder
| MP165.1
| MP166.1
| MP122.2
| no sample
| D166
| no sample
| D167
| no sample
| D131
| no sample
| 100pb ladder
|
| Expected size (pb)
|
| 2 957
| 2 996
| 2 957
|
| 2942
|
| 914
|
| 900
|
| Measured size (pb)
|
| 3 000
| 3 000
| 3 000
|
| 2500
|
| 2500
|
| 950
|
|
|
=> Following a mistake, the right E0430(D166) and E0432(D167) digestion, have not been purified.
So we need to repeat the same digestion experiment tomorrow morning.
Construction of: promotor-rbs-LasR-dbl ter
Number of colonies
| Ligation name
| Description
| Antibio
| Number Colonies observed
| Fluorescence
| Comments
|
| Ligations
|
| L153
| D123(BV) - D165(BI)
J23100 - rbs-LasR-Double terminator
| Amp
| 768
| No
| OK
|
| L154
| D103(BV) - D165(FV)
J23101 - rbs-LasR - Double terminator
| Amp
| 236
| No
| OK
|
| Controls
|
| C1
| D123(BV)
| Amp
| 23
| No
| OK
|
| C2
| D103(FV)
| Amp
| 144
| No
| OK
|
| Positive Control
| pUC19
| Amp
| 2264 (efficiency 4,5.10^8)
| No
| OK
|
PCR Screening
Protocol
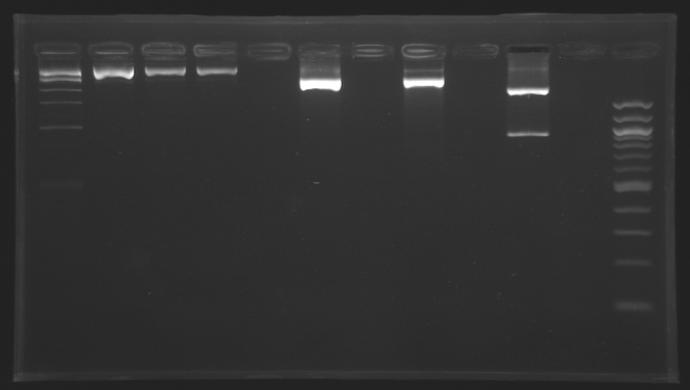
 Ligations results J23100+D165 and J23101+D165
| Well
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
| 13
| 14
| 15
| 16
| 17
|
| Sample
| L153
| 1kb ladder
| L154
|
| Clone
| .1
| .2
| .3
| .4
| .5
| .6
| .7
| .8
|
| .1
| .2
| .3
| .4
| .5
| .6
| .7
| .8
|
| Expected size
| 1194
|
| 1194
|
| Measured size
| 1200
|
| 1200
|
Minipreps
| Miniprep Name
| Ligation name
| Antibio
| Biobricks
| Description
|
| MP169.1
| L153.1
| Amp
|     
| J23100 - rbs-LasR-Double terminator
|
| MP169.2
| L153.2
|
| MP170.1
| L154.1
|     
| J23101 - rbs-LasR - Double terminator
|
| MP170.2
| L154.2
|
Promoter characterization plasmids
Ligation
Our ligations from yesterday didn't work. The positive control for transformation worked.
Digestion
We had a problem with a gel extraction so we have to make again the digestions from yesterday, Other digestions made:
Protocol Digestion
| Digestion name
| Plasmid
| Description
| Miniprep used
| Enzymes
| Concentration after gel extraction
|
| D179
| MP3.4
| B0015 (double terminator B0010-B0012) - BV
| 4
| SpeI and PstI
| 9
|
| D180
| MP101.1
| promoter J23101- BV
| 1
| SpeI and PstI
| 7
|
| D181
| MP104.2
| PTet (TetR repressible promoter) - FV
| 1
| EcoRI and XbaI
| 1
|
| D182
| MP114.1
| TetR - BI
| 1
| XbaI and PstI
| 10
|
| D183
| MP119.3
| pBad promoter - BI
| 1
| XbaI and PstI
| 0
|
| D184
| MP143.1
| gfp generator - FI
| 2
| EcoRI and SpeI
| 13
|
| D185
| MP163.1
| B0032 RBS - BV
| 2
| SpeI and PstI
| 21
|
D179
D180
D181
D182
D183
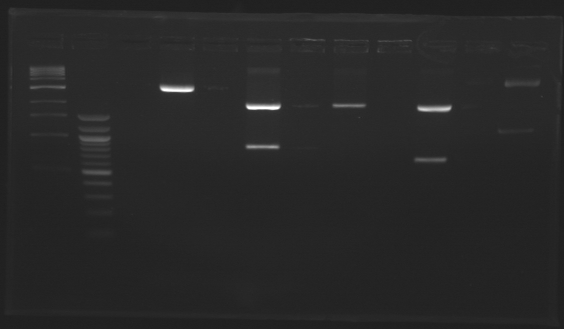
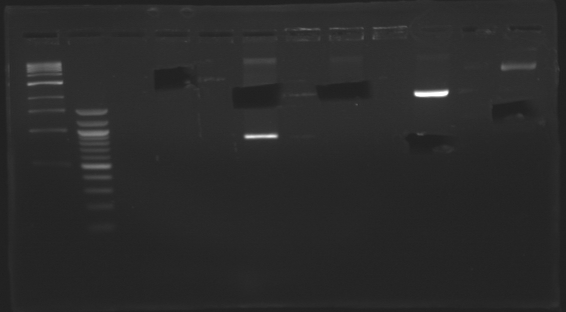
D184
D185
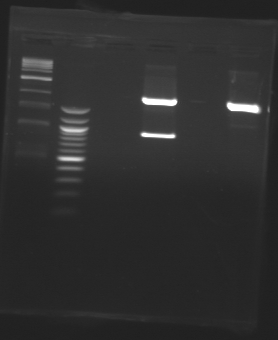
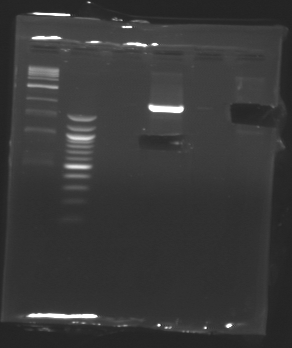
Sequencing
- We received results of our sequencing from COCHIN
- We succeed for FlgA promoter
- FlgB and flhB don't match with our expected sequences. We decided to cut our Miniprep product with other enzymes to check our sequence.
Digestion
Protocol
- D176 : pFlgB digested with ApoI
- D177 : pFlgB digested with NruI
- D178 : pFlhB digested with BstAPI
Screening
Gel 1
| Well
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
|
| Sample
| 1kb ladder
| D176.1
| D176.2
| D176.3
| D176.4
| D177.1
| D177.2
| 100pb ladder
|
| Expected size
|
| 2970
194
48
| 3212
|
| Measured size
|
| 2900
200
/
| Plamsid not digested
|
|
Gel 2
| Well
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
|
| Sample
| nothing
| D177.3
| D177.4
| D178.1
| D178.2
| D178.3
| D178.4
| 1kb ladder
|
| Expected size
|
| 3212
| 3213
|
| Measured size
|
| Plamsid not digested
|
|
Conclusion => the sequence of pFlgB and pFlhB are not good we will tried to isolate pFlhB again.
|
 "
"