Team:Paris/August 19
From 2008.igem.org
(Difference between revisions)
(→Screening of the cloning of OmpR*, EnvZ* and FlhDC+promotor) |
(→Ligation) |
||
| (35 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
=Screening of the cloning of OmpR*, EnvZ* and FlhDC+promotor= | =Screening of the cloning of OmpR*, EnvZ* and FlhDC+promotor= | ||
| + | ==Electrophoresis== | ||
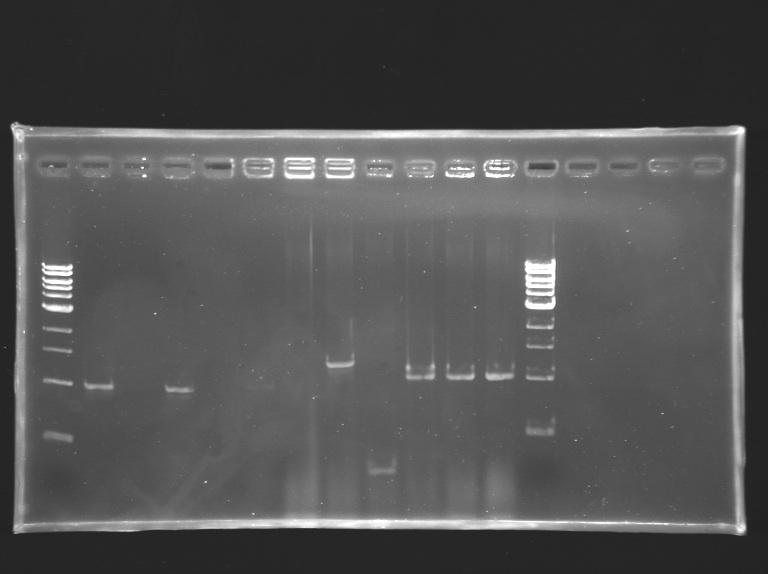
| + | [[Image:KR000188.jpg|thumb|Screening of the cloning of OmpR*, EnvZ* and FlhDC+promotor]] | ||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
|'''well n°''' | |'''well n°''' | ||
| Line 20: | Line 22: | ||
|- | |- | ||
|'''sample''' | |'''sample''' | ||
| - | |1 kb DNA ladder | + | |1 kb <br>DNA ladder |
| - | | | + | |control +<br> pSB3K3 <br>(S158) |
| - | | | + | |control -<br>no<br>template |
| + | |colspan="3"|OmpR* | ||
| + | |colspan="3"|EnvZ* | ||
| + | |colspan="3"|FlhDC+promotor | ||
| + | |1 kb<br>DNA ladder | ||
| + | |- | ||
| + | |'''ligation/clone''' | ||
| + | | | ||
| + | | | ||
| + | | | ||
|L133.1 | |L133.1 | ||
|L133.2 | |L133.2 | ||
| Line 32: | Line 43: | ||
|L132.2 | |L132.2 | ||
|L132.3 | |L132.3 | ||
| - | | | + | | |
|- | |- | ||
|'''expected size''' | |'''expected size''' | ||
| | | | ||
| - | | | + | |316 bp |
|0 kb | |0 kb | ||
|colspan="3"|about 1 kb | |colspan="3"|about 1 kb | ||
|colspan="3"|about 1,5 kb | |colspan="3"|about 1,5 kb | ||
| - | |colspan="3"| | + | |colspan="3"|1210 bp |
| | | | ||
|- | |- | ||
|'''measured size''' | |'''measured size''' | ||
| | | | ||
| - | |1 kb | + | |style="background: #ff6d73"|1 kb |
| - | |0 kb | + | |style="background: #ff6d73"|0 kb |
| - | |1 kb | + | |style="background: #cbff7B"|1 kb |
| - | |0 kb | + | |style="background: #ff6d73"|0 kb |
| - | |0 kb | + | |style="background: #ff6d73"|0 kb |
| - | |0 kb | + | |style="background: #ff6d73"|0 kb |
| - | |1,4 kb | + | |style="background: #cbff7B"|1,4 kb |
| - | |<0,5 kb | + | |style="background: #ff6d73"|<0,5 kb |
| - | |1 kb | + | |style="background: #ff6d73"|1 kb |
| - | |1 kb | + | |style="background: #ff6d73"|1 kb |
| - | |1 kb | + | |style="background: #ff6d73"|1 kb |
| | | | ||
|} | |} | ||
| - | + | ||
| - | The clones L133.2 and L133.3 didn't grow up in LB+ampicillin: they seem not to have the plasmid, as revealed by PCR. | + | |
| + | ==Minipreps and glycerol stock== | ||
| + | |||
| + | *The clones L133.2 and L133.3 didn't grow up in LB+ampicillin: they seem not to have the plasmid, as revealed by PCR. | ||
| + | *Minipreps and Glycerol stocks were made for the clones L133.1 and L134.2. | ||
| + | |||
| + | {|border="1" style="text-align: center" | ||
| + | |'''Miniprep''' | ||
| + | |'''Glycerol Stock | ||
| + | |'''Ligation''' | ||
| + | |'''Name''' | ||
| + | |- | ||
| + | |MP155 | ||
| + | |S154 | ||
| + | |L133.1 | ||
| + | |OmpR* | ||
| + | |- | ||
| + | |MP156 | ||
| + | |S155 | ||
| + | |L134.2 | ||
| + | |EnvZ* | ||
| + | |} | ||
| + | |||
| + | *==>Minipreps of L133.1 and L134.2 will be sequenced. | ||
=Screening of the cloning of E0240 and FlhDC+promotor= | =Screening of the cloning of E0240 and FlhDC+promotor= | ||
| Line 69: | Line 103: | ||
|'''DNA cloned''' | |'''DNA cloned''' | ||
|'''vector''' | |'''vector''' | ||
| - | |'''size of the fragment amplified by VF & VR''' | + | |'''expected size of the fragment amplified by VF & VR''' |
| + | |'''mesured size''' | ||
|- | |- | ||
|S159.1 | |S159.1 | ||
| Line 77: | Line 112: | ||
|pSB3K3 | |pSB3K3 | ||
|1192 bp | |1192 bp | ||
| + | |1,5 kb<br>'''1,1 kb'''<br>0,6 kb | ||
|- | |- | ||
|S161.1 | |S161.1 | ||
| Line 83: | Line 119: | ||
|FlhDC+promotor | |FlhDC+promotor | ||
|pSB1A2 | |pSB1A2 | ||
| - | | | + | |1403 bp |
| + | |'''1,4'''<br>0,4 kb<br>0,3 kb | ||
|} | |} | ||
| Line 95: | Line 132: | ||
*Spreading of 200 µL in a LB agar plate containing the appropriate antibiotic | *Spreading of 200 µL in a LB agar plate containing the appropriate antibiotic | ||
*Incubation overnight at 37°C | *Incubation overnight at 37°C | ||
| + | |||
| + | |||
| + | |||
| + | =Promoter characterization plasmids= | ||
| + | |||
| + | ==Ligation== | ||
| + | |||
| + | [[Team:Paris/Notebook/Protocols#Ligation |Protocol]] | ||
| + | |||
| + | {|border="1" style="text-align: center" | ||
| + | |'''Ligation name''' | ||
| + | |'''Vector digestion''' | ||
| + | |'''Vector description''' | ||
| + | |'''Vector vol. (µL)''' | ||
| + | |'''Insert digestion''' | ||
| + | |'''Insert description''' | ||
| + | |'''Insert vol. (µL)''' | ||
| + | |'''Product description''' | ||
| + | |'''Antibiotic''' | ||
| + | |- | ||
| + | |L155 | ||
| + | |D164 | ||
| + | |J23101 promoter | ||
| + | |10 | ||
| + | |D163 | ||
| + | |gfp generator | ||
| + | |2 | ||
| + | |J23101 promoter-gfp generator | ||
| + | |Amp | ||
| + | |- | ||
| + | |L156 | ||
| + | |D161 | ||
| + | |pTet promoter | ||
| + | |1 | ||
| + | |D163 | ||
| + | |gfp generator | ||
| + | |4 | ||
| + | |pTet promoter-gfp generator | ||
| + | |Kana | ||
| + | |- | ||
| + | |TL156 | ||
| + | |D161 | ||
| + | |1 | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |Vector autoligation control | ||
| + | |Kana | ||
| + | |- | ||
| + | |L157 | ||
| + | |D125.2 | ||
| + | |B0015 | ||
| + | |3 | ||
| + | |D162 | ||
| + | |tetR | ||
| + | |4 | ||
| + | |tetR-B0015 | ||
| + | |Amp | ||
| + | |- | ||
| + | |TL157 | ||
| + | |D125.2 | ||
| + | |3 | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |Vector autoligation control | ||
| + | |Amp | ||
| + | |} | ||
| + | |||
| + | ==Transformation== | ||
| + | [[Team:Paris/Notebook/Protocols#Transformation |Protocol]] | ||
| + | |||
| + | These transformations were made during the day at 16°C | ||
| + | |||
| + | ==Digestion== | ||
| + | |||
| + | ===Measurement of concentration of minipreps=== | ||
| + | to be modified | ||
| + | [[Team:Paris/Notebook/Protocols#Concentration of the Miniprep|standard protocol]] | ||
| + | |||
| + | {| border="1" style="text-align: center" | ||
| + | |'''Plasmid''' | ||
| + | |'''Miniprep''' | ||
| + | |'''Concentration (µg/mL)''' | ||
| + | |'''ratio 260/280''' | ||
| + | |- | ||
| + | |MP3 | ||
| + | |3 | ||
| + | |38 | ||
| + | |1.72 | ||
| + | |- | ||
| + | |MP3 | ||
| + | |4 | ||
| + | |30 | ||
| + | |1.70 | ||
| + | |- | ||
| + | |MP101 | ||
| + | |1 | ||
| + | |333 | ||
| + | |1.68 | ||
| + | |- | ||
| + | |MP101 | ||
| + | |2 | ||
| + | |416 | ||
| + | |1.66 | ||
| + | |- | ||
| + | |MP101 | ||
| + | |4 | ||
| + | |200 | ||
| + | |1.74 | ||
| + | |- | ||
| + | |MP104 | ||
| + | |1 | ||
| + | |145 | ||
| + | |1.63 | ||
| + | |- | ||
| + | |MP104 | ||
| + | |3 | ||
| + | |147 | ||
| + | |1.29 | ||
| + | |- | ||
| + | |MP104 | ||
| + | |4 | ||
| + | |51 | ||
| + | |1.66 | ||
| + | |- | ||
| + | |MP114 | ||
| + | |1 | ||
| + | |173 | ||
| + | |1.75 | ||
| + | |- | ||
| + | |MP114 | ||
| + | |2 | ||
| + | |263 | ||
| + | |1.43 | ||
| + | |- | ||
| + | |MP119 | ||
| + | |1 | ||
| + | |42 | ||
| + | |1.62 | ||
| + | |- | ||
| + | |MP119 | ||
| + | |2 | ||
| + | |26 | ||
| + | |1.56 | ||
| + | |- | ||
| + | |MP119 | ||
| + | |3 | ||
| + | |39 | ||
| + | |1.64 | ||
| + | |- | ||
| + | |MP143 | ||
| + | |1 | ||
| + | |138 | ||
| + | |1.69 | ||
| + | |- | ||
| + | |MP143 | ||
| + | |2 | ||
| + | |150 | ||
| + | |1.61 | ||
| + | |- | ||
| + | |MP163 | ||
| + | |1 | ||
| + | |80 | ||
| + | |1.61 | ||
| + | |- | ||
| + | |MP163 | ||
| + | |2 | ||
| + | |79 | ||
| + | |1.69 | ||
| + | |} | ||
| + | |||
| + | ===Digestion=== | ||
| + | [[Team:Paris/Notebook/Protocols#Digestion|Protocol Digestion]] | ||
| + | |||
| + | {| border="1" style="text-align: center" | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |'''Miniprep used''' | ||
| + | |'''Enzymes''' | ||
| + | |- | ||
| + | |MP118.1 | ||
| + | |B0015 (double terminator B0010-B0012) - FV | ||
| + | |4 | ||
| + | |EcoRI and XbaI | ||
| + | |- | ||
| + | |MP119 | ||
| + | |promoter J23101 - BV | ||
| + | |1 | ||
| + | |SpeI and PstI | ||
| + | |- | ||
| + | |MP104 | ||
| + | |PTet (Tet promoter) - BV | ||
| + | |1 | ||
| + | |SpeI and PstI | ||
| + | |- | ||
| + | |MP114 | ||
| + | |TetR - FI | ||
| + | |1 | ||
| + | |EcoRI and SpeI | ||
| + | |- | ||
| + | |MP143 | ||
| + | |gfp generator - BI | ||
| + | |2 | ||
| + | |SpeI and PstI | ||
| + | |} | ||
| + | |||
| + | We had a problem with a gel and we lost these digestions. | ||
Latest revision as of 20:11, 4 September 2008
Screening of the cloning of OmpR*, EnvZ* and FlhDC+promotorElectrophoresis
Minipreps and glycerol stock
Screening of the cloning of E0240 and FlhDC+promotorSpreading the clones in order to obtain single colonies
The PCR screening of the transformants L139 and L142 of august 15th revealed several bands for a given clone including one band appearing at the right size.
In order to check these 2 hypothesis and to isolate (if it is possible) the right clone (containing the plasmid with the insert). We decided to spread the "clone" in question in a LB plate in order to carry out a PCR screening on single colonies.
Promoter characterization plasmidsLigation
TransformationThese transformations were made during the day at 16°C DigestionMeasurement of concentration of miniprepsto be modified standard protocol
Digestion
We had a problem with a gel and we lost these digestions. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"