Team:Paris/August 21
From 2008.igem.org
(Difference between revisions)
AnaJimenez (Talk | contribs) (→Ligations) |
(→Ligations) |
||
| (17 intermediate revisions not shown) | |||
| Line 17: | Line 17: | ||
==Result== | ==Result== | ||
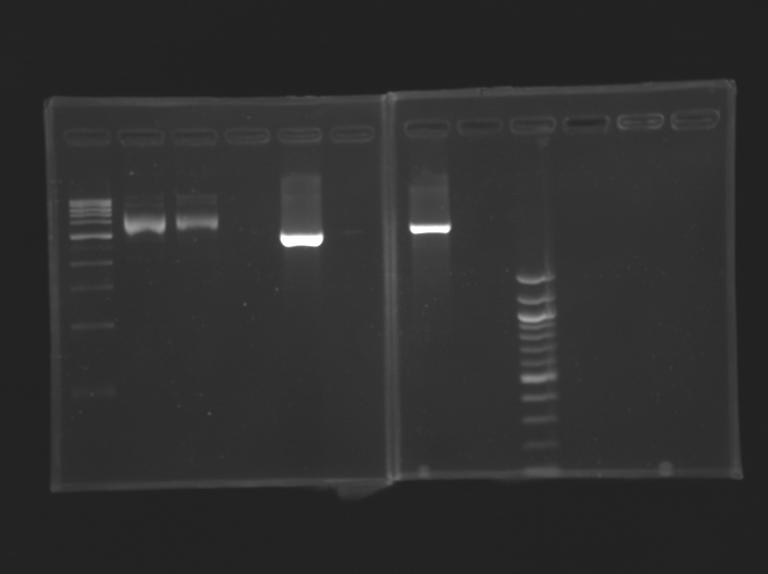
| - | [[Image:KR00019b.jpg| thumb| Vérification of pFlhB] | + | [[Image:KR00019b.jpg| thumb| Vérification of pFlhB]] |
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| Line 70: | Line 70: | ||
| - | + | =Construction for FIFO= | |
| - | + | ||
| - | + | ||
| - | = | + | |
Aim : Construction of pFlgA - YFP tripart (+/- LVA) ''' "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432)''' [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | Aim : Construction of pFlgA - YFP tripart (+/- LVA) ''' "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432)''' [[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| Line 124: | Line 121: | ||
|- | |- | ||
|'''Sample''' | |'''Sample''' | ||
| - | |1kb ladder | + | |1kb<br>ladder |
|MP165.1 | |MP165.1 | ||
|MP166.1 | |MP166.1 | ||
| Line 132: | Line 129: | ||
|D167 | |D167 | ||
|no sample | |no sample | ||
| - | |100pb ladder | + | |100pb<br>ladder |
|colspan="3"|no sample | |colspan="3"|no sample | ||
|- | |- | ||
| Line 150: | Line 147: | ||
|style="background: #cbff7B"| 3 000 | |style="background: #cbff7B"| 3 000 | ||
| | | | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|2 800 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| | | | ||
| + | |style="background: #cbff7B"|3 000 | ||
| + | |colspan="5"| | ||
|} | |} | ||
| Line 211: | Line 206: | ||
|} | |} | ||
| - | |||
| - | We obtained | + | =Screening of the cloning of E0240 and FlhDC+promotor= |
| - | + | ||
| + | We obtained colonies isolated with the dilution 1/100; 13 clones were analysed by PCR | ||
==PCR screening== | ==PCR screening== | ||
| Line 260: | Line 255: | ||
|- | |- | ||
|'''sample''' | |'''sample''' | ||
| - | |1 kb DNA ladder | + | |1 kb<br> DNA ladder |
| - | | | + | |control + |
| - | | | + | |control - |
|colspan="13"|S159.1 | |colspan="13"|S159.1 | ||
| - | |100 bp DNA ladder | + | |100 bp<br>DNA ladder |
|- | |- | ||
|'''colonie n°''' | |'''colonie n°''' | ||
| Line 323: | Line 318: | ||
|- | |- | ||
|'''sample''' | |'''sample''' | ||
| - | |1 kb DNA ladder | + | |1 kb<br>DNA ladder |
| - | | | + | |control + |
| - | | | + | |control - |
|colspan="13"|S161.1 | |colspan="13"|S161.1 | ||
| - | |100 bp DNA ladder | + | |100 bp<br>DNA ladder |
|- | |- | ||
|'''colonie n°''' | |'''colonie n°''' | ||
| Line 359: | Line 354: | ||
| | | | ||
| | | | ||
| - | |colspan="13"|0,3 kb | + | |style="background: #ff6d73" colspan="13"|0,3 kb |
| | | | ||
|} | |} | ||
<br> | <br> | ||
| - | '''Results''': | + | '''Results''': |
| - | *The clone of E0240 (S159.1) always have several bands amplified by PCR. It might contain different plasmids. | + | *The clone of E0240 (S159.1) always have several bands amplified by PCR.<br>It might contain different plasmids. |
| - | *The clone of FlhDC+promotor (S161.1) don't have the correct size band. It also doesn't have the insert in the plasmid. | + | *The clone of FlhDC+promotor (S161.1) don't have the correct size band. <br>It also doesn't have the insert in the plasmid. |
| - | = | + | =Construction for synchronization= |
| - | == | + | ==Ligations== |
[[Team:Paris/Notebook/Protocols#Ligation |Protocol]] | [[Team:Paris/Notebook/Protocols#Ligation |Protocol]] | ||
| Line 386: | Line 381: | ||
|D131 (BI) | |D131 (BI) | ||
|2.89 | |2.89 | ||
| + | |- | ||
| + | |control L158 | ||
| + | |rbs-TetR + gfp tripart <br> [[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]][[Image:Part_icon_rbs.png]][[Image:Part_icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| + | |D110 (BV) | ||
| + | |2 | ||
| + | | | ||
| + | | | ||
|- | |- | ||
|L159 | |L159 | ||
| - | |rbs-lasI | + | |rbs-lasI <br>[[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]] |
|D125 (FV) | |D125 (FV) | ||
|2.08 | |2.08 | ||
|D109 (FI) | |D109 (FI) | ||
|1.15 | |1.15 | ||
| + | |- | ||
| + | |control L159 | ||
| + | |rbs-lasI <br>[[Image:Part_icon_rbs.png]][[Image:Icon_coding.png]] | ||
| + | |D125 (FV) | ||
| + | |2.08 | ||
| + | | | ||
| + | | | ||
|} | |} | ||
| - | |||
=Promoter characterization plasmids= | =Promoter characterization plasmids= | ||
| - | == | + | ==Ligations from digestions from 20th== |
| - | + | ||
| - | + | ||
| - | + | '''Top 10 cells were used''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[Team:Paris/Notebook/Protocols#Transformation |Protocol]] | ||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| Line 495: | Line 416: | ||
|'''Vector digestion''' | |'''Vector digestion''' | ||
|'''Vector description''' | |'''Vector description''' | ||
| + | |'''Vector conc. ug/mL''' | ||
|'''Vector volume''' | |'''Vector volume''' | ||
|'''Insert digestion''' | |'''Insert digestion''' | ||
|'''Insert description''' | |'''Insert description''' | ||
| + | |'''Insert conc. ug/mL''' | ||
|'''Insert volume''' | |'''Insert volume''' | ||
|'''Product description''' | |'''Product description''' | ||
| Line 505: | Line 428: | ||
|D164 | |D164 | ||
|J23101 promoter | |J23101 promoter | ||
| - | | | + | |3 |
| + | |16 | ||
|D163 | |D163 | ||
|gfp generator | |gfp generator | ||
| - | | | + | |14 |
| + | |8 | ||
|J23101 promoter-gfp generator | |J23101 promoter-gfp generator | ||
|Amp | |Amp | ||
| Line 515: | Line 440: | ||
|D161 | |D161 | ||
|pTet promoter | |pTet promoter | ||
| - | |1 | + | |39 |
| + | |1.25 | ||
|D163 | |D163 | ||
|gfp generator | |gfp generator | ||
| - | | | + | |14 |
| + | |5 | ||
|pTet promoter-gfp generator | |pTet promoter-gfp generator | ||
|Kana | |Kana | ||
|- | |- | ||
| - | | | + | |Control L156 |
|D161 | |D161 | ||
| | | | ||
| - | |1 | + | | |
| + | |1.25 | ||
| + | |none | ||
| | | | ||
| | | | ||
| Line 535: | Line 464: | ||
|D125.2 | |D125.2 | ||
|B0015 | |B0015 | ||
| + | |15 | ||
|3 | |3 | ||
|D162 | |D162 | ||
| - | |||
|tetR | |tetR | ||
| + | |11 | ||
| + | |4 | ||
|tetR-B0015 | |tetR-B0015 | ||
| + | |Kana | ||
| + | |- | ||
| + | |Control L157 | ||
| + | |D125.2 | ||
| + | | | ||
| + | | | ||
| + | |3 | ||
| + | |none | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |Vector autoligation control | ||
| + | |Kana | ||
| + | |- | ||
| + | |L166 | ||
| + | |D185 | ||
| + | |RBS B0032 | ||
| + | |21 | ||
| + | |2 | ||
| + | |D182 | ||
| + | |tetR | ||
| + | |10 | ||
| + | |6.5 | ||
| + | |RBS B0032 - tetR | ||
|Amp | |Amp | ||
|- | |- | ||
| + | |Control L166 | ||
| + | |D185 | ||
| | | | ||
| - | |||
| | | | ||
| + | |2 | ||
| + | |none | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |Vector autoligation control | ||
| + | |Amp | ||
| + | |- | ||
| + | |L167 | ||
| + | |D181 | ||
| + | |pTet | ||
| + | |1 | ||
| + | |14 | ||
| + | |D184 | ||
| + | |gfp generator | ||
| + | |14 | ||
|3 | |3 | ||
| + | |gfp generator - pTet | ||
| + | |Amp | ||
| + | |- | ||
| + | |Control L167 | ||
| + | |D181 | ||
| + | |pTet | ||
| + | | | ||
| + | |14 | ||
| + | |none | ||
| | | | ||
| | | | ||
Latest revision as of 14:29, 6 September 2008
Cloning of FlhB promoterProtocol
Resuspension of 1 colony E.coli K12 strain MG 1655 in 100µl of water.
1µl of dNTP
ResultFile:KR00019b.jpg Vérification of pFlhB
Construction for FIFOAim : Construction of pFlgA - YFP tripart (+/- LVA) "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432) DigestionDigestion
Gel Extraction
Measurement of the concentration of D166 & D167 purifiedProtocol (it's same that for Miniprep)
Ligation
Screening of the cloning of E0240 and FlhDC+promotorWe obtained colonies isolated with the dilution 1/100; 13 clones were analysed by PCR PCR screeningreaction mixture (25 µL)
PCR screening programm
Electrophoresis
Results: *The clone of E0240 (S159.1) always have several bands amplified by PCR. Construction for synchronizationLigationsPromoter characterization plasmidsLigations from digestions from 20thTop 10 cells were used
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"