Team:Paris/August 22
From 2008.igem.org
(Difference between revisions)
AnaJimenez (Talk | contribs) (→Transformation of ligations from August 20th) |
(→Transformation of the ligations we did yesterday) |
||
| (15 intermediate revisions not shown) | |||
| Line 73: | Line 73: | ||
|'''measured size''' | |'''measured size''' | ||
| | | | ||
| - | |'''1 kb'''<br>3 kb | + | |style="background: #ff6d73" |'''1 kb'''<br>3 kb |
| | | | ||
| - | |0,3 kb<br>1,8 kb | + | |style="background: #ff6d73" |0,3 kb<br>1,8 kb |
| - | |0,3 kb<br>1,8 kb | + | |style="background: #cbff7B"| 0,3 kb<br>1,8 kb |
| - | |0,9 kb<br>2 kb | + | |style="background: #ff6d73" |0,9 kb<br>2 kb |
| - | |2 kb | + | |style="background: #ff6d73" |2 kb |
| - | |2 kb | + | |style="background: #ff6d73" |2 kb |
| | | | ||
|} | |} | ||
| - | + | ||
| + | '''Results''': The clones tested didn't have the insert. | ||
| + | |||
='''Construction of pFlgA - GFP Generator'''= | ='''Construction of pFlgA - GFP Generator'''= | ||
| Line 180: | Line 182: | ||
| - | | - | ||
|} | |} | ||
| + | |||
='''Construction for FIFO'''= | ='''Construction for FIFO'''= | ||
| Line 202: | Line 205: | ||
|- | |- | ||
|L161 | |L161 | ||
| - | |D167 (FV) + | + | |D167 (FV) + D132 (FI)<br>pFlgA - YFP tripart (LVA+) |
|Amp | |Amp | ||
|- | |- | ||
| Line 303: | Line 306: | ||
| - | =Promoter characterization plasmids= | + | ='''Promoter characterization plasmids'''= |
==Transformation of ligations from August 20th== | ==Transformation of ligations from August 20th== | ||
| - | |||
| - | |||
| - | |||
| - | |||
[[Team:Paris/Notebook/Protocols#Transformation |Protocol]] | [[Team:Paris/Notebook/Protocols#Transformation |Protocol]] | ||
| - | + | '''Top 10 cells were used''' | |
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| Line 318: | Line 317: | ||
|'''Vector digestion''' | |'''Vector digestion''' | ||
|'''Vector description''' | |'''Vector description''' | ||
| - | |'''Vector | + | |'''C° Vector (µg/mL)''' |
| + | |'''V. Vector (µl)''' | ||
|'''Insert digestion''' | |'''Insert digestion''' | ||
|'''Insert description''' | |'''Insert description''' | ||
| - | |'''Insert | + | |'''C° Insert (µg/mL)''' |
| + | |'''V. Insert (µl)''' | ||
|'''Product description''' | |'''Product description''' | ||
|'''Antibiotic''' | |'''Antibiotic''' | ||
| Line 328: | Line 329: | ||
|D164 | |D164 | ||
|J23101 promoter | |J23101 promoter | ||
| - | | | + | |3 |
| + | |16 | ||
|D163 | |D163 | ||
|gfp generator | |gfp generator | ||
| - | | | + | |14 |
| + | |8 | ||
|J23101 promoter-gfp generator | |J23101 promoter-gfp generator | ||
|Amp | |Amp | ||
| Line 338: | Line 341: | ||
|D161 | |D161 | ||
|pTet promoter | |pTet promoter | ||
| - | |1 | + | |39 |
| + | |1.25 | ||
|D163 | |D163 | ||
|gfp generator | |gfp generator | ||
| - | | | + | |14 |
| + | |5 | ||
|pTet promoter-gfp generator | |pTet promoter-gfp generator | ||
|Kana | |Kana | ||
|- | |- | ||
| - | | | + | |Control L156 |
|D161 | |D161 | ||
| | | | ||
| - | |1 | + | | |
| + | |1.25 | ||
| + | |none | ||
| | | | ||
| | | | ||
| Line 358: | Line 365: | ||
|D125.2 | |D125.2 | ||
|B0015 | |B0015 | ||
| + | |15 | ||
|3 | |3 | ||
|D162 | |D162 | ||
| - | |||
|tetR | |tetR | ||
| + | |11 | ||
| + | |4 | ||
|tetR-B0015 | |tetR-B0015 | ||
|Kana | |Kana | ||
| Line 367: | Line 376: | ||
|Control L157 | |Control L157 | ||
|D125.2 | |D125.2 | ||
| + | | | ||
| | | | ||
|3 | |3 | ||
| + | |none | ||
| | | | ||
| | | | ||
| Line 378: | Line 389: | ||
|D185 | |D185 | ||
|RBS B0032 | |RBS B0032 | ||
| + | |21 | ||
|2 | |2 | ||
|D182 | |D182 | ||
|tetR | |tetR | ||
| + | |10 | ||
|6.5 | |6.5 | ||
|RBS B0032 - tetR | |RBS B0032 - tetR | ||
| Line 387: | Line 400: | ||
|Control L166 | |Control L166 | ||
|D185 | |D185 | ||
| - | | | + | | |
| + | | | ||
|2 | |2 | ||
|none | |none | ||
| + | | | ||
| | | | ||
| | | | ||
| Line 398: | Line 413: | ||
|D181 | |D181 | ||
|pTet | |pTet | ||
| + | |1 | ||
|14 | |14 | ||
|D184 | |D184 | ||
|gfp generator | |gfp generator | ||
| + | |14 | ||
|3 | |3 | ||
|gfp generator - pTet | |gfp generator - pTet | ||
| Line 408: | Line 425: | ||
|D181 | |D181 | ||
|pTet | |pTet | ||
| + | | | ||
|14 | |14 | ||
|none | |none | ||
| + | | | ||
| | | | ||
| | | | ||
Latest revision as of 00:54, 21 September 2008
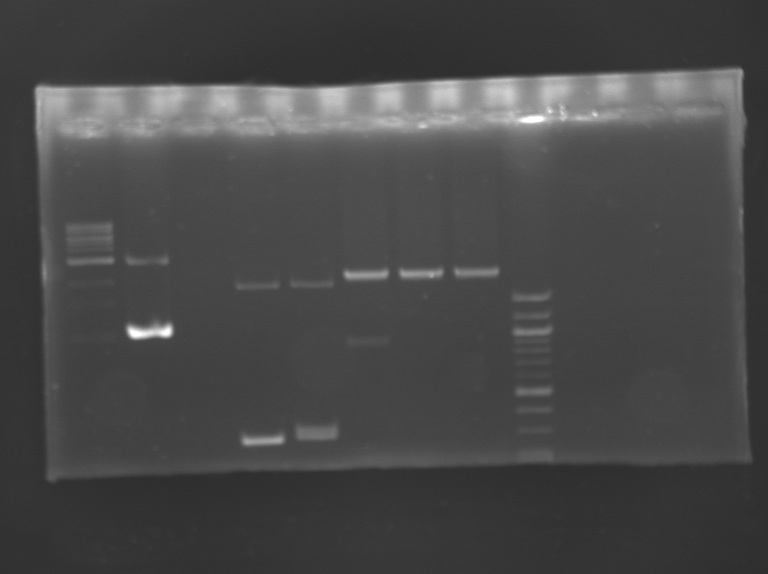
Analysis of the transformant of FlhDC+promotor
PCRPCR screening programm
Digestiontotal volume reaction (30 µL)
Incubation 2h55 at 37°C and then 20 min at 65°C. Electrophoresis
Results: The clones tested didn't have the insert.
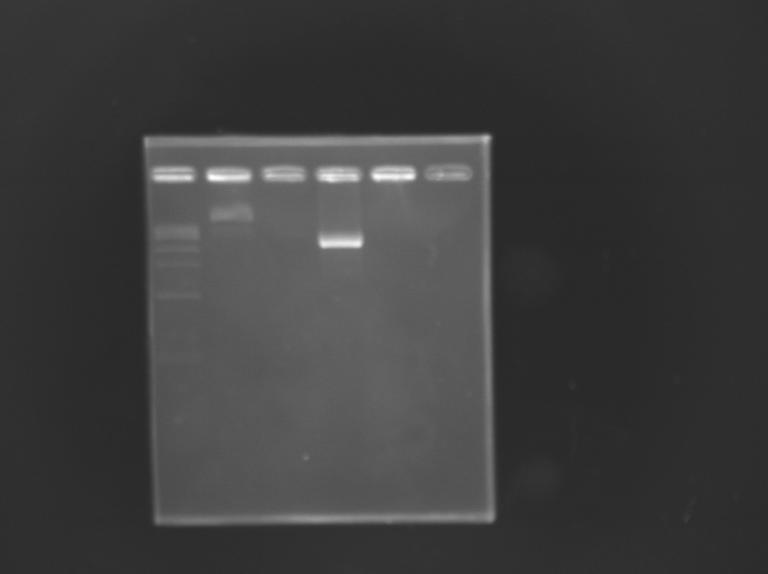
Construction of pFlgA - GFP GeneratorAim : Construction of "pFlgA-RBS-GFP-dbl ter" (pFlgA-E0240) DigestionDigestion
Gel Extraction
Measurement of the concentration of D168 purifiedProtocol (it's same that for Miniprep)
Ligation
Construction for FIFOAim : Construction of pFlgA - YFP tripart (+/- LVA) "pFlgA-RBS-YFP-dbl ter" (pFlgA-E0430/E0432) Transformation of the ligations we did yesterday
Construction for synchronizationTransformation of the ligations we did yesterday
LigationTransformation
Promoter characterization plasmidsTransformation of ligations from August 20thTop 10 cells were used
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"