Team:Paris/August 25
From 2008.igem.org
Construction for SynchronizationTransformation of the ligations we did yesterday
Construction of pFlgA - GFP GeneratorAim : Construction of "pFlgA-RBS-GFP-dbl ter" (pFlgA-E0240)
Results of the transformation we did the day before yesterday
=> Need to screen to know which clones we can use for the of pFlgA promotor characterization. Cloning of EnvZ*The sequencing of EnvZ* previously cloned, revealed a loss of about 300 bp. EnvZ* contains indeed an EcoRI restriction site within its sequence. So we can't use this enzyme during the cloning. Digestion
Reaction mixture
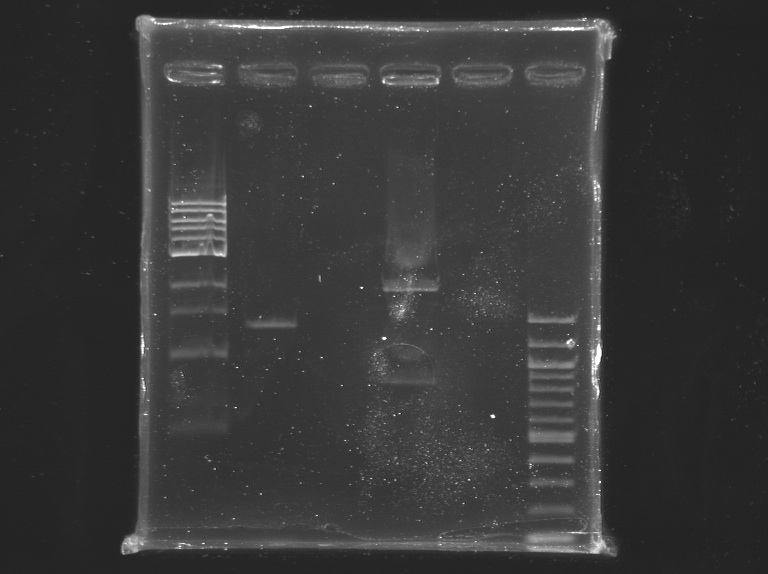
Incubation at 37°C during 2H25, and then ~20 min at 65°C Electrophoresis1% agarose gel
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"