|
← Yesterday ↓ Calendar ↑Tomorrow →
Extraction of EnvZ* and OmpR* from E. coli genome
Name of the PCR
| Name
| What's in ?
| Template DNA
| Oligo F
| Oligo R
|
| PCR 147
| EnvZ*
| S 120
| O 126
| O 127
|
| PCR 148
| OmpR*
| S 119
| O 138
| O 139
|
| T 147
| nothing
| no template
| O 126
| O 127
|
| T 148
| nothing
| no template
| O 138
| O 139
|
Protocol
Protocol
- 10 µL Phusion HF Buffer 5X
- 2.5 µL Oligo F 10 mM
- 2.5 µL Oligo R 10 mM
- 1 µL dNTP
- 1 µL Template DNA
- 33 µL H2O
PCR Programme
- 98°C 30s Initial denaturation
- CYCLE 30X
- 98°C 10s Denaturation
- 60°C 30s Annealing
- 72°C 45s Elongation
- END OF CYCLE
- 72°C 5min Terminal Elongation
Resuts
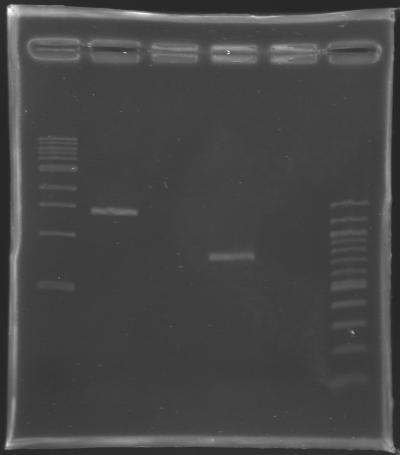 Results of the cloning of EnvZ* and OmpR* Electrophoresis settings
- Gel 1% agar
- 10µL Ladder 1kb
- 10µL Ladder 100bp
- 4µL DNA + 2µL Loading Blue
Results of the electrophoresis
| Name
| Gene
| Well
| Expected size
| Measured size
|
| PCR 147
| EnvZ*
| 2
| 1412 bp
| 1400 bp
|
| T 147
| nothing
| 3
| nothing
| nothing
|
| PCR 148
| OmpR*
| 4
| 779 bp
| 800 bp
|
| T 148
| nothing
| 5
| nothing
| nothing
|
Conclusion : All the PCR worked perfectly well !
Cleaning of the PCR products
Standard protocol
The cleaned PCR products are stored in the freezer overnight.
PCR
PCR1
- pFlhDC (O111-F / O113-R)
- Gene FlhDC (O134-F / O131-R)
- pFlgB (O102-F / O103-R)
- pFlhB (O108-F / O109-R)
Program: Gradient
Denaturation :
98°C-5'
Cycling 1 (3X) :
98°C-10"
Gradient 61°C +/-10°C - 30
72°C-30"
Cycling 2 (25X) :
98°C-10"
72°C-30"
Elongation :
72°C-5'
Vf=20µL
H20=13,4µL
Buffer5X=4µL
dNTP=0,4µL
O1=1µL
O2=1µL
Phusion=0,2µL
PCR2
- pFliA (rbs) (O145-F / O144-R)
- pFliA (O145-F / O146-R)
- pFliA +Gene FliA (O145-F / O151-R)
Program: promoter
Denaturation :
98°C-5'
Cycling 1 (3X) :
98°C-10"
55°C - 30
72°C-30"
Cycling 2 (30X) :
98°C-10"
65°C-30"
72°C-30"
Elongation :
72°C-5'
Vf=50µL
H20=33,5µL
Buffer5X=10µL
dNTP=1µL
O1=2,5µL
O2=2,5µL
Phusion=0,5µL
PCR3
- PCRFliA1 (O143-F / O152-R)
- PCRFliA2 (O153-F / O150-R)
- PCRFliA3 (O148-F / O150-R)
Program: PCRFliA1
Denaturation :
98°C-5'
Cycling 1 (30X) :
98°C-10"
Gradient 66°C +/-6°C - 25
72°C-20"
Elongation :
72°C-5'
Program: PCRFliA2
Denaturation :
98°C-5'
Cycling 1 (3X) :
98°C-10"
Gradient 61°C +/-10°C - 25
72°C-20"
Cycling 2 (30X) :
98°C-10"
72°C-30"
Elongation :
72°C-5'
Program: PCRFliA3
Denaturation :
98°C-30'
Cycling 1 (2X) :
98°C-10"
72°C-30"
Cycling 2 (5X) :
98°C-10"
98°C->72°C low decreasing 1.0°C/s
72°C-30"
Elongation :
72°C-5'
| Name
| Promotor
| Well
| Expected size
| Measured size
|
| PCRFliA1
| Upstream part of FliA
| 2-27
| 197 bp
| ~ 200 bp
|
| PCRFliA2
| Downstream part of FliA
| 28-54
| 0 bp
| 0 bp
|
| PCR
| pFliA+rbs
| 55-56
| 325 bp
| 0 bp
|
| PCR
| PFliA
| 57-58
| 310 bp
| 0 bp
|
| PCR
| pFliA+Gene FliA
| 59-60
| 1100 bp
| 0 bp
|
Cloning of EnvZ*
Concentration measurement by Biophotometer
| Name
| Digestion n°
| [DNA] in µg/mL
| A260/A280
|
| EnvZ*
| D159
| 10 µg/mL
| 1.35
|
| pSB1A2
| D116
| 17 µg/mL
| 1.02
|
Ligation
|
| control
| insert / vector mass ratio
|
|
|
| 1,8 / 1
| 2,4 / 1
| 3,1 / 1
|
| D159 EnvZ* (µL)
| 0
| 6
| 8
| 8
|
| D116 pSB1A2 (µL)
| 2
| 2
| 2
| 1,5
|
| 10X T4 ligase (µL)
| 2
| 2
| 2
| 2
|
| T4 DNA ligase (µL)
| 1
| 1
| 1
| 1
|
| water (µL)
| 15
| 9
| 7
| 7,5
|
- 5 hours at room temperature
- transformation of TOP10 competent cells with 5 µL of ligation product
- spreading on LB plates + ampicilline and incubation overnight at 37°C
Miniprep and stock glycerolof New Biobrick
| Miniprep
| Strain
| Name
| Description
|
| MP1.1
| S1.1
|
| 
RBS
|
| MP1.2
| S1.2
|
| MP1.1
| S1.1
|
|    
RBS+ ECFP+ LVA+ term
|
| MP1.2
| S1.2
|
| MP1.1
| S1.1
|
|    
RBS+ YFP+ LVA- term
|
| MP1.2
| S1.2
|
| MP1.1
| S1.1
|
|    
RBS+ YFP LVA+ term
|
| MP1.2
| S1.2
|
| MP1.1
| S1.1
|
|    
RBS+ ECFP LVA- term
|
| MP1.2
| S1.2
|
Construction of pFlhB - mRFP Tripart (LVA+)
Aim : Construction of "pFlhB-RBS-mRFP-dbl ter" (pFlhB-I732078)     
We can only do the construction with mRFP Tripart (LVA+) because the stable strain with the Biobricks I732011 (mRFP Tripart LVA-) don't to growth.
Digestion
Digestion
Protocol Digestion
| Name
| Template DNA
| Description
| Vol MP (µl)
| Vol H2O (µl)
| Enzymes
|
| D187
| MP168.1
| RBS - mRFP - term (FV)
| 9.00
| 15.7
| EcoRI and XbaI
|
Gel Verification
Protocol
| Well
| 1
| 2
| 3
| 4
| 5
| 6
|
| Sample
| 100 pb ladder
| MP168.1
| no sample
| D187
| no sample
|
| Expected size (pb)
|
|
|
|
|
|
| Measured size (pb)
|
|
|
|
|
|
Screening of the cloning of pFlgA-GFP Generator
Electrophoresis
| well n°
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
|
| sample
| 1kb ladder
| MP172.6
| MP143.1
| L161.1
| L161.2
| L161.3
| L161.4
| L161.5
| L161.6
|
| expected size (pb)
|
|
| 876
| 1 126
|
| measured size
|
|
|
|
|
|
|
|
|
|
Minipreps and glycerol stock
| Miniprep
| Glycerol Stock
| Ligation
| Name
|
| MP1
| S1
| L164
| FlgA-rbs-GFP-dbl ter
|
|
 "
"

