Team:Paris/August 5
From 2008.igem.org
(Difference between revisions)
(→Electrophoresis Purification of PCR) |
|||
| (12 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Paris/Calendar_Links|August 4|August 6}} | |
| - | '' | + | == '''Amplification of Promotors of interest (FliA, FliL, FlgA, FlgB, FlhB, FlhDC)'''== |
| - | + | ''We performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.'' | |
| - | + | ||
| + | === '''PCR Protocol''' === | ||
| - | + | * '''Preparation of the templates''' :--> Resuspend of 1 colony of MG1655 in 100µl of water. | |
| - | |||
| - | |||
| - | |||
| + | * '''List of Oligos''' : | ||
{| | {| | ||
| - | |align="center"|Name | + | |- style="background: #649CD7;" |
| - | |align="center"|Promotor | + | |- style="background: #649CD7; text-align: center;" |
| - | |align="center"|Gel | + | |width=5%| Number |
| - | |align="center"|Band | + | |width=10%| Name |
| - | |align="center"|Expected size | + | |width=40%| Sequence |
| - | |align="center"|Measured size | + | |width=5%| Length |
| + | |width=35%| Comments | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" |O100 | ||
| + | | FlgA-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGAGCATATCTCCTCCGCAGGTATCAAAAT | ||
| + | | 58 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O101 | ||
| + | | FlgA-R | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTAACAGTATCGCGATGATCGCCACGCTACGT | ||
| + | | 58 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O102 | ||
| + | | FlgB-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGACAGTATCGCGATGATCGCCACGCTACG | ||
| + | | 58 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O103 | ||
| + | | FlgB-R | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTAAGCATATCTCCTCCGCAGGTATCAAAATT | ||
| + | | 58 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O108 | ||
| + | | FlhB-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGCCACGTCATATCAGGCGGTCTGATAAGG | ||
| + | | 58 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O109 | ||
| + | | FlhB-R | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTAGTTTTGTCGTCGCTCTCGTCAGACACGTC | ||
| + | | 58 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O111 | ||
| + | | FlhDC-Total-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGTCATTTTTGCTTGCTAGCGTACGGAAAA | ||
| + | | 58 | ||
| + | |Amplify the both OmpR binding site | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O113 | ||
| + | | FlhDC(nu)-R | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTACAGAATAACCAACTTTATTTTTATG | ||
| + | | 54 | ||
| + | |Don't amplify the natural rbs of FlhD (only promoter) | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O124 | ||
| + | | FliL-F | ||
| + | | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGCAGCGAGAGGCTGTTGGTATTAATGACT | ||
| + | | 58 | ||
| + | | | ||
| + | |- style="background: #dddddd;" | ||
| + | | style="background: #D4E2EF;" | O125 | ||
| + | | FliL-R | ||
| + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTACCAGCGATGAAATACTTGCCATGCGATTT | ||
| + | | 58 | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | * '''Preparation of PCR mix''' : | ||
| + | ''For each samples,'' | ||
| + | |||
| + | |||
| + | 1 µl dNTP | ||
| + | <br>10 µl Buffer Phusion 5x | ||
| + | <br>2,5 µl Oligo_F | ||
| + | <br>2,5 µl Oligo_R | ||
| + | <br>1µl template | ||
| + | <br>1 µl Phusion | ||
| + | <br>50 µl qsp H2O (33µl) | ||
| + | |||
| + | |||
| + | * Program PCR: Annealing 55°C - Time élongation 1'30" - Number cycle : 29 | ||
| + | |||
| + | |||
| + | |||
| + | === Electrophoresis Purification of PCR=== | ||
| + | |||
| + | ''When the PCR cycles were finished,'' | ||
| + | |||
| + | '''conditions :''' | ||
| + | |||
| + | * 10µl of ladder 1 kb (unlike 100 pb) | ||
| + | * 2 x 30µl of PCR products added with 10µl of loading Dye 6x | ||
| + | * migration ~30min at 100W on a '''1,5% agarose gel'''. | ||
| + | |||
| + | |||
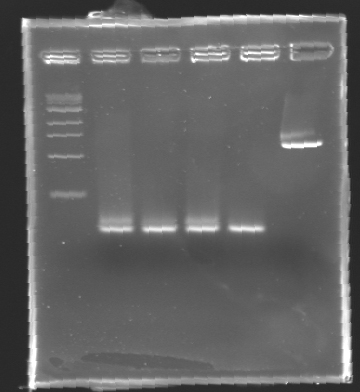
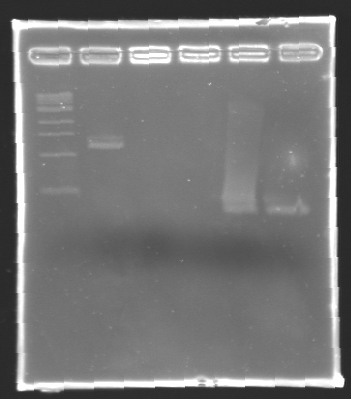
| + | '''Results of electrophoresis''' | ||
| + | |||
| + | <br>gel 1 [[Image:KR000102.jpg| gel 1|150px]] | ||
| + | gel 2 [[Image:KR000104.jpg|gel 2|145px]] | ||
| + | |||
| + | {|- border="1" | ||
| + | |align="center"|'''Name''' | ||
| + | |align="center"|'''Promotor''' | ||
| + | |align="center"|'''Gel''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
|- | |- | ||
|align="center"|PCR_124 | |align="center"|PCR_124 | ||
|align="center"|pFlgA | |align="center"|pFlgA | ||
| - | |align="center"| | + | |align="center"|1 |
| - | |align="center"| | + | |align="center"|2-3 |
| - | | | + | |style="background: #cbff7B"|<center>261 pb</center> |
| - | |align="center"| | + | |align="center"|300 pb |
|- | |- | ||
|align="center"|PCR_125 | |align="center"|PCR_125 | ||
|align="center"|pFlgB | |align="center"|pFlgB | ||
| - | |align="center"| | + | |align="center"|1 |
| - | |align="center"| | + | |align="center"|4-5 |
| - | | | + | |style="background: #cbff7B"|<center>261 pb</center> |
| - | |align="center"| | + | |align="center"|300 pb |
|- | |- | ||
|align="center"|PCR_126 | |align="center"|PCR_126 | ||
|align="center"|pFlhB | |align="center"|pFlhB | ||
| - | |align="center"| | + | |align="center"|2 |
| - | |align="center"| | + | |align="center"|5-6 |
| - | | | + | |style="background: #cbff7B"|<center>260 pb</center> |
| - | |align="center"| | + | |align="center"|300 pb |
|- | |- | ||
|align="center"|PCR_127 | |align="center"|PCR_127 | ||
|align="center"|pFlhDC | |align="center"|pFlhDC | ||
| - | |align="center"| | + | |align="center"|1 & 2 |
| - | |align="center"| | + | |align="center"|7 & 2 |
| - | | | + | |style="background: #ff6d73"|<center>446 pb</center> |
| - | |align="center"| | + | |align="center"|1,000 pb |
|} | |} | ||
| - | ==> '''Remark :''' | + | |
| + | ==> '''Remark :''' for PCR the negative control (templates = water) can be check on the gel n°2, on the band 3-4 | ||
| + | |||
| + | |||
| + | ==> '''Conclusion:''' for the promotors '''FlgA, FlgB, FlhB''' we observe the size expected. | ||
| + | <br>We need to repeat the experiments for the promotors '''FlhDC'''. | ||
| + | |||
| + | |||
| + | * After electrophoresis, the bands corresponding to the right amplification were excised and purified using the QIAquick DNA Gel Extraction Kit by "Maurice (QIAcube)". | ||
| + | * Elution in 50 µL of buffer EB. | ||
| + | * Store at -20°C | ||
| + | |||
| + | == Culture of J61002== | ||
| + | |||
| + | * 3 x 5ml LB with Ampicilin, cloning of one clone of J612002 | ||
| + | * Culture O/N at 37°C, on 225 rmp. | ||
| + | * will be use to do minipreps | ||
Latest revision as of 15:17, 6 August 2008
Amplification of Promotors of interest (FliA, FliL, FlgA, FlgB, FlhB, FlhDC)We performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR Protocol
For each samples,
Electrophoresis Purification of PCRWhen the PCR cycles were finished, conditions :
Culture of J61002
|
 "
"