|
← Yesterday ↓ Calendar ↑Tomorrow →
We performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR Protocol
For each samples,
- 1 µl dNTP
- 10 µl Buffer Phusion 5x
- 2,5 µl VR2 (O18)
- 2,5 µl VF (O19)
- 1 µl Phusion
- 50 µl qsp H2O (33µl)
Electrophoresis
When the PCR cycles were finished,
- Add 10 µL of 6X loading dye were added
- The samples were then loaded (2 x 30 µL per sample) on a 1,5% agarose gel
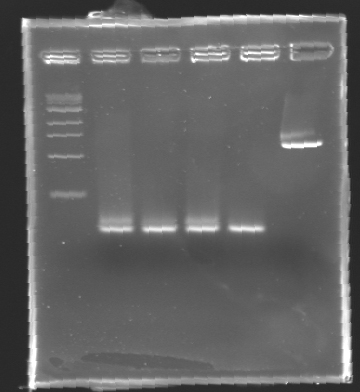
After electrophoresis, the bands corresponding to MP 100 and MP 120 were excised and purified using the QIAquick DNA Gel Extraction Kit (QIAGEN). The elution was made in 50 µL of water. Because the intensity of the band corresponding to MP 120 was very low, we only continued with MP 100. MP 100 was digested by EcoRI & SpeI (Forward Insert) or by XbaI & PstI (D100 : Backward Insert).

DNA Digestion
Digestion reaction (total volume : 50 µL)
- 25 µL DNA
- 5 µL buffer 2 (10X)
- 2 µL enzyme 1
- 2 µL enzyme 2
- 0.5 µL BSA (100X)
- 15,5 µL water
- Incubate 2 hours at 37°C
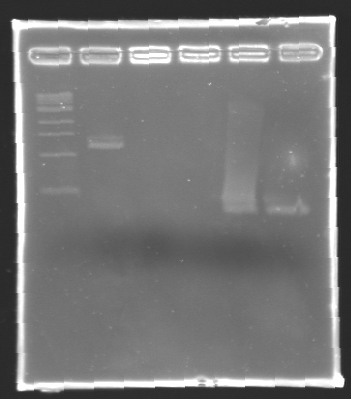
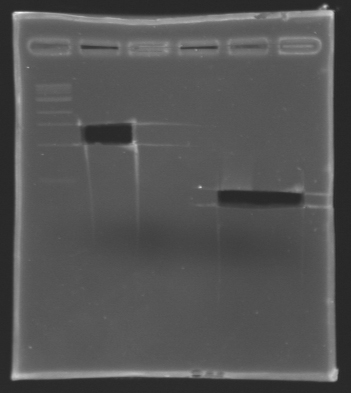
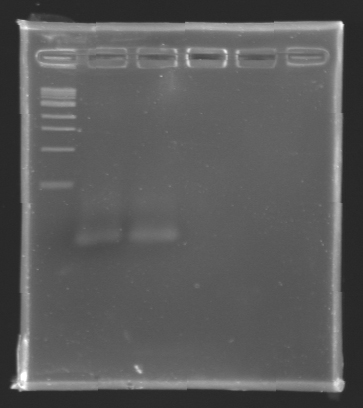
- The samples (2 x 30 µL per sample) were then analysed by electrophoresis.
Electrophoresis (bis)
The sequence of MP 100 (B0034) digested by EcoRI & SpeI (35 bp) or XbaI & PstI (34 bp) was too short and we didn't manage to visualise it on the gel.
Conclusion : small parts like B0034 can't be cloned as an insert.
|
 "
"






