Team:Paris/July 24
From 2008.igem.org
(Difference between revisions)
(→Results of digestions : Electrophoresis) |
(→MiniPreps) |
||
| (14 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
== MiniPreps == | == MiniPreps == | ||
| - | *The | + | *The Promega MiniPreps protocol has been used on all the clones cultivated on the 23th. |
{| border="1" | {| border="1" | ||
| Line 11: | Line 11: | ||
|align="center"|'''Description''' | |align="center"|'''Description''' | ||
|- | |- | ||
| - | |align="center"|MP116 | + | |align="center"|MP116 |
| - | |align="center"|J23100 | + | |align="center"|[http://partsregistry.org/Part:BBa_J23100 J23100] |
| - | |align="center"| | + | |align="center"|Strong constitutive promoter in J61002 |
|- | |- | ||
| - | |align="center"|MP117 | + | |align="center"|MP117 |
| - | |align="center"|J23107 | + | |align="center"|[http://partsregistry.org/Part:BBa_J23107 J23107] |
| - | |align="center"| | + | |align="center"|Medium constitutive promoter in J61002 |
|- | |- | ||
| - | |align="center"|MP118 | + | |align="center"|MP118 |
| - | |align="center"|B0015 | + | |align="center"|[http://partsregistry.org/Part:BBa_B0015 B0015] |
| - | |align="center"| | + | |align="center"|Double terminator |
|- | |- | ||
| - | |align="center"|MP119 | + | |align="center"|MP119 |
| - | |align="center"|I0500 | + | |align="center"|[http://partsregistry.org/Part:BBa_I0500 I0500] |
| - | |align="center"| | + | |align="center"|AraC pBAD |
|- | |- | ||
| - | |align="center"|MP120 | + | |align="center"|MP120 |
| - | |align="center"|B0030 | + | |align="center"|[http://partsregistry.org/Part:BBa_B0030 B0030] |
| - | |align="center"| | + | |align="center"|Strong RBS (Efficiency = 0,6) |
|- | |- | ||
| - | |align="center"|MP121 | + | |align="center"|MP121 |
| - | |align="center"|E0422 | + | |align="center"|[http://partsregistry.org/Part:BBa_E0422 E0422] |
| - | |align="center"| | + | |align="center"|ECFP (RBS+LVA+Term) |
|- | |- | ||
| - | |align="center"|MP122 | + | |align="center"| MP122 |
| - | |align="center"|E0840 | + | |align="center"|[http://partsregistry.org/Part:BBa_E0840 E0840] |
| - | |align="center"| | + | |align="center"|gfp tri-part; strong rbs |
| + | | | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
== Digestion == | == Digestion == | ||
| - | |||
===Digestion Mix=== | ===Digestion Mix=== | ||
| - | 10µl of Miniprep ( | + | 10µl of Miniprep (26 aug.) <br> |
12.5µl of water<br> | 12.5µl of water<br> | ||
2.5µl of Buffer N°2<br> | 2.5µl of Buffer N°2<br> | ||
| Line 55: | Line 52: | ||
1µl of enzyme 1<br> | 1µl of enzyme 1<br> | ||
1µl of enzyme 2<br> | 1µl of enzyme 2<br> | ||
| + | |||
| + | * Incubation 1h at 37°C with the first enzyme | ||
| + | * Add the second enzyme | ||
| + | * Incubation 1h at 37°C with the second enzyme | ||
| + | * Store on ice | ||
| + | * Revelation of the digestion by electrophoresis on agarose gel | ||
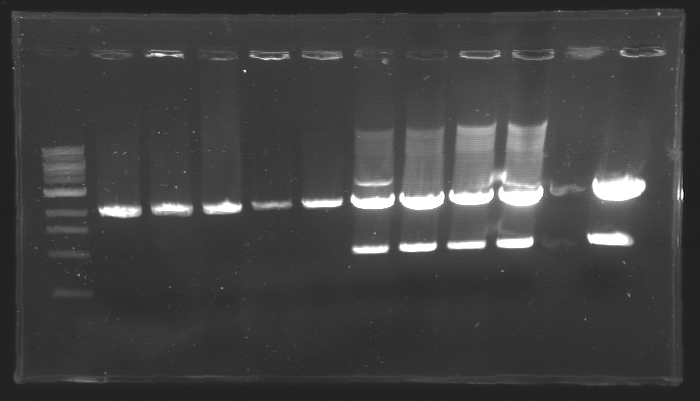
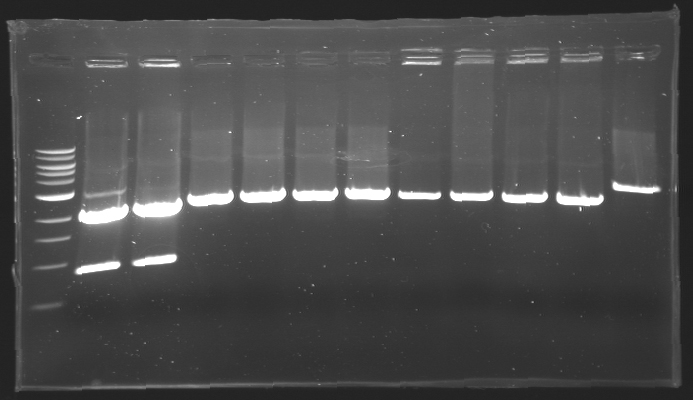
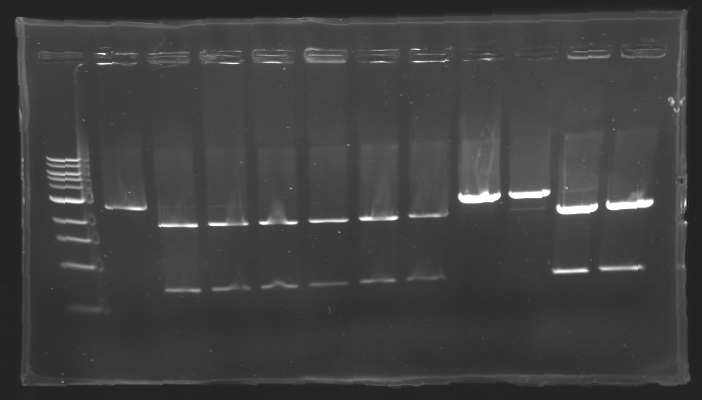
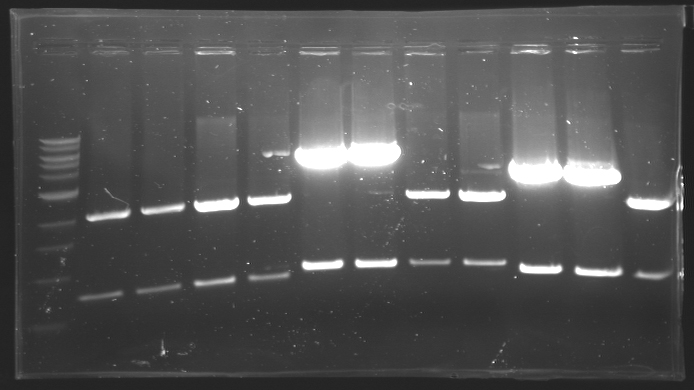
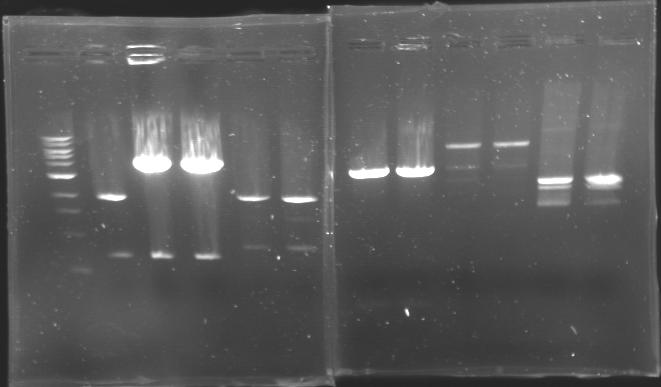
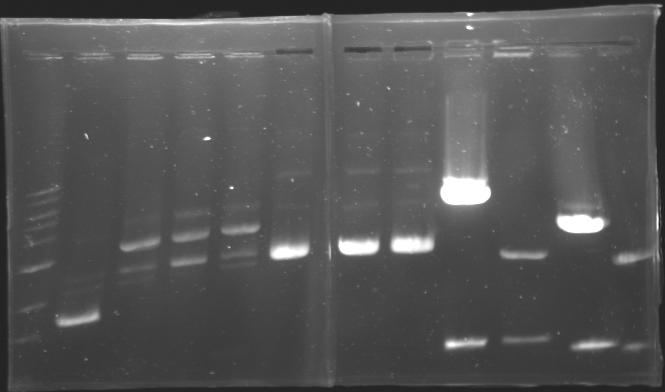
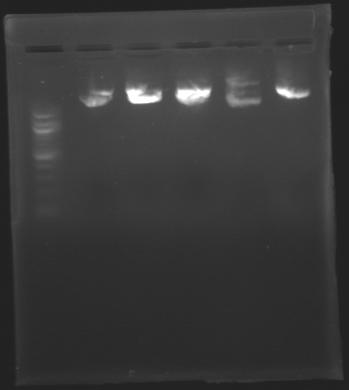
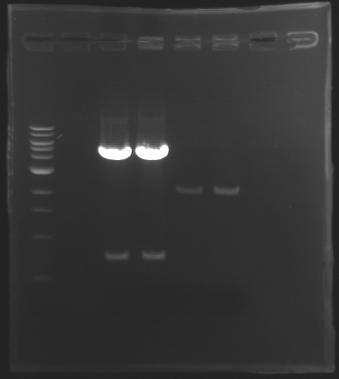
=== Results of digestions : Electrophoresis === | === Results of digestions : Electrophoresis === | ||
| + | |||
| + | conditions : | ||
| + | * 10µl of ladder 1 kb (except for gel n°7 : 100 pb) | ||
| + | * 30µl of digestion added with 5µl of loading Dye 6x | ||
| + | * migration ~30min at 100W | ||
| + | * Gel 1, 2, 3, 4, 5, 6, 8 = '''0.8%''' | ||
| + | * Gel 7 = '''1,2%''' | ||
| Line 210: | Line 220: | ||
! rowspan="2"| XbaI | ! rowspan="2"| XbaI | ||
! rowspan="2"| PstI | ! rowspan="2"| PstI | ||
| - | ! rowspan="2"| | + | ! rowspan="2"| BI |
! rowspan="2"| 2057 pb | ! rowspan="2"| 2057 pb | ||
! rowspan="2"| 707 pb | ! rowspan="2"| 707 pb | ||
| Line 225: | Line 235: | ||
| align=center|EcoRI | | align=center|EcoRI | ||
| align=center|SpeI | | align=center|SpeI | ||
| - | | align=center| | + | | align=center|FI |
| align=center| 2056 pb | | align=center| 2056 pb | ||
| align=center| 708 pb | | align=center| 708 pb | ||
| Line 249: | Line 259: | ||
! rowspan="2"| XbaI | ! rowspan="2"| XbaI | ||
! rowspan="2"| PstI | ! rowspan="2"| PstI | ||
| - | ! rowspan="2"| | + | ! rowspan="2"| BI |
! rowspan="2"| 2057 pb | ! rowspan="2"| 2057 pb | ||
! rowspan="2"| 725 pb | ! rowspan="2"| 725 pb | ||
| Line 264: | Line 274: | ||
| align=center|EcoRI | | align=center|EcoRI | ||
| align=center|SpeI | | align=center|SpeI | ||
| - | | align=center| | + | | align=center|FI |
| align=center| 2756 pb | | align=center| 2756 pb | ||
| align=center| 726 pb | | align=center| 726 pb | ||
| Line 511: | Line 521: | ||
|align=center| 6-8 | |align=center| 6-8 | ||
|} | |} | ||
| + | |||
| + | |||
| + | ==> Conclusion: Most of the digestion have succeed | ||
| + | |||
| + | == Extraction of the DNA == | ||
| + | |||
| + | |||
| + | * Cutting of the parts of interest, for all the digestion that have migrated on the gels | ||
| + | * Store of all the piece of gel O/N at -20°C. | ||
Latest revision as of 14:39, 31 July 2008
|
MiniPreps
DigestionDigestion Mix10µl of Miniprep (26 aug.)
Results of digestions : Electrophoresisconditions :
Extraction of the DNA
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"