From 2008.igem.org
(Difference between revisions)
|
|
| Line 178: |
Line 178: |
| | | | |
| | == Transformations == | | == Transformations == |
| | + | |
| | + | === Protocol === |
| | + | |
| | + | |
| | + | use of TOP10 Chemically competent cells |
| | + | |
| | + | * Defroze competent cells on ice during 5' |
| | + | * Add 5µl of DNA Ligation in 50µL of competent bacterias (or 1µL for the positive control puc19) |
| | + | * Incubate 30' on ice |
| | + | * Heat-shock the cells during 30" at 42°C without shaking |
| | + | * Put 2' on ice |
| | + | * Add 250µL of pre-warmed SOC medium (4°C) |
| | + | * Incubate 1h at 37°C under shaking (225rpm) |
| | + | * Spin at 5.000rpm during 30" |
| | + | * Remove 150µL of supernatant |
| | + | * Resuspent the pellet in the 150µL left |
| | + | * Spread on adequated plates |
| | + | * Incubate O/N at 37°C |
Revision as of 10:28, 5 August 2008
|
← Yesterday ↓ Calendar ↑Tomorrow →
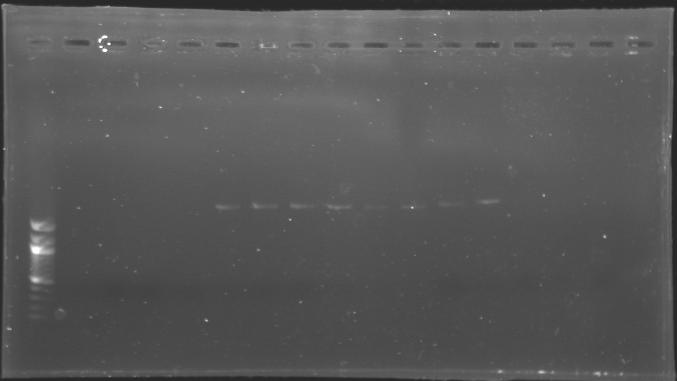
DNA digestion and purification
Mix digestion
for each reaction (total volume : 50 µL)
- 20 µL of DNA (MiniPrep product)
- 2 µL of enzyme 1
- 2 µL of enzyme 2
- 5 µL of buffer 2 (10X)
- 0,5 µL of BSA
- 20,5 µL water
Protocol
Each reaction was :
- Incubate 2 hours at 37°C, then 10 minutes at 60-65°C (to inactivate the enzymes).
- Add 10 µL of loading dye (6X) to each of the 50 µL of digestion product.
- Run the whole samples in a 1,5% agarose gel (about 30 minutes at 100 W ; 2 x 30 µL per sample ; 30 µL per well).
- Excise the bands of interest from the gel and the DNA was purified using the QIAquick DNA Gel Extraction kit (QIAGEN).
- The elution of DNA was performed using 50 µL of water (after 10 minutes of incubation at 37°C).
==> Unfortunately, there were not enough columms, so we took some columms from the QIAGEN MiniPrep kit, hoping that it will work with the QIAquick DNA Gel Extraction kit. Some of the samples were too voluminous, so we separated them into two tubes.
Each of the samples was then analysed by a 1,5% agarose gel:
- 2 µL of DNA
- 3 µL of water
- 1 µL of 6X loading dye
The ladder used was the 100 bp ladder from New England Biolabs.
| Name
| BioBrick
| Tube N°
| Enz 1
| Enz 2
| Obs
| Exp Size
| Mea Size
| Conc (ng/µl)
| Band
|
| D101
| B0034
| 1
| EcoRI
| XbaI
| FV
| 2076 pb
|
|
| 10
|
| D102
| B0034
| 1
| SpeI
| PstI
| BV
| 2077 pb
|
|
| 11
|
| D105
| R0079
| 1
| SpeI
| PstI
| BV
| 2222 pb
|
|
| 13
|
| D106
| R0040
| 1
| SpeI
| PstI
| BV
| 2119 pb
|
|
| 12
|
| D108
| S03154
| 1
| XbaI
| PstI
| BI
| 707 pb
|
|
| 14
|
| D111
| S03879
| 1
| XbaI
| PstI
| BI
| 725 pb
|
|
| 15
|
| D115
| C0179
| 2
| EcoRI
| SpeI
| FI
| 746 pb
|
|
| 4 & 5
|
| D116
| C0179
| 2
| XbaI
| PstI
| BI
| 745 pb
|
|
| 2 & 3
|
| D117
| E0030
| 1
| EcoRI
| SpeI
| FI
| 746 pb
|
|
| 16
|
| D128
| B0030
| 2
| EcoRI
| XbaI
| FV
| 2079 pb
|
|
| 6 & 7
|
| D129
| B0030
| 2
| SpeI
| PstI
| BV
| 2080 pb
|
|
| 8 & 9
|
==> Conclusion :The amount of DNA loaded was too low and the DNA ladder used was the wrong one. So this experiment has to be reconducted tomorrow.
Transformations
Protocol
use of TOP10 Chemically competent cells
- Defroze competent cells on ice during 5'
- Add 5µl of DNA Ligation in 50µL of competent bacterias (or 1µL for the positive control puc19)
- Incubate 30' on ice
- Heat-shock the cells during 30" at 42°C without shaking
- Put 2' on ice
- Add 250µL of pre-warmed SOC medium (4°C)
- Incubate 1h at 37°C under shaking (225rpm)
- Spin at 5.000rpm during 30"
- Remove 150µL of supernatant
- Resuspent the pellet in the 150µL left
- Spread on adequated plates
- Incubate O/N at 37°C
|
|
 "
"