Team:Paris/August 6
From 2008.igem.org
|
Cloning of flagella gene promotors into J61002 plasmidWe have to digest (by EcoRI and SpeI) the PCR products of yesterday (amplification of pFlgA (MP124), pFlgB (MP125), pFlhB (MP126) and pFlhDC (MP127) in order to clone them into the J61002 plasmid (that we must extract and then digest by EcoRI and SpeI).
Plasmid extractionThe J61002 plasmid was extracted from overnight bacteria culture using the QIAspin Miniprep Kit (QIAGEN) by QIACube.
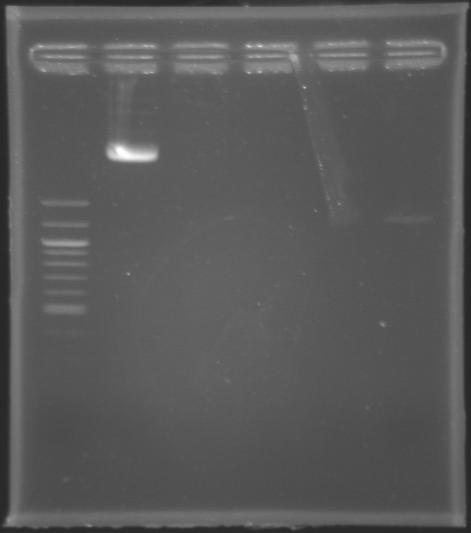
Quantification of DNAIn order to quantify the DNA contained either in the Miniprep product of MP123 or in the PCR products of MP124, MP125, MP126 and MP127 previously purifed yesterday by the QIAquick Gel Extraction Kit, we carried out : By electrophoresis
==>Results : The MP123 plasmid is clearly visible but the PCR products (purified by QIAquick Gel Extraction) aren't. There might be a problem during the purification step using the QIAquick Gel Extraction. Whatever, we still go on with the digestion. By Spectrophotometer
==> Conclusion :we took the MP123 n°2 to do digestions
Digestion
For each sample (MP123 and PCR products of MP124, MP125, MP126 and MP127):
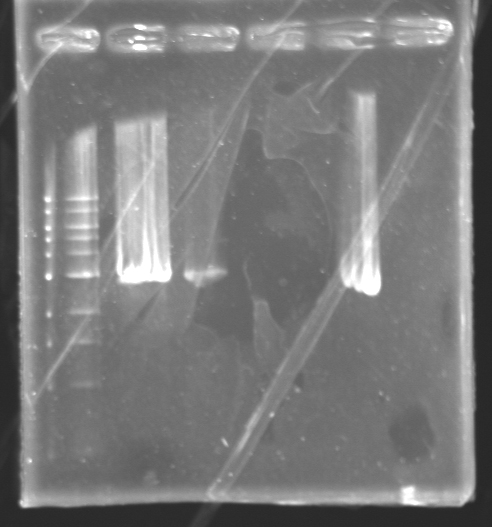
Electrophoresis of the digested J61002 plasmid
Analysis of the purified digested DNA:Excision, dissolution and purification of the band of interest using the Wizard SV Gel and PCR Clean-Up System (Promega).
=> Concentration : +/- 12ng/4µL -> 3ng/µL LigationProtocolFor each sample,
List of ligations
Cloning of pflhDCYesterday the isolation of pflhDC did not work : the PCR product measured more than 1.5 kb. We checked the primers, they are well designed but they contain a sequence similarity with other sequences in E.coli K12. To amplify more precisely the promoter, we decided to do a PCR with gradient. Protocol
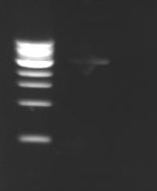
PCR ProgramPROMOTE2 * LID : 105 °C *1. T: 95°C 5min *2. T: 95°C 1min *3. T: 55°C 30s ~>G: 5°C *4. T: 72°C 1min30 *5. GO TO: 2 REP: 29 *6. T: 72°C 5min *7. HOLD: 10°C ResultsOn the left side, the temperature was 50°C, in the center 55°C and on the right 60°C. We can't see anything on this electrophoresis (except the ladder) ! Remarks :
The return of PCRs for amplification of promotersAs our results were not very encouraging, we started again a PCR to amplify:
ProtocolWe used the taq polymerase, following a typical protocol. ProgramThe PCR Program was a new version of PROMOTE2 : PROMOTE2 * LID : 105 °C *1. T: 95°C 5min *2. T: 95°C 1min *3. T: 60°C 30s ~>G: 5°C *4. T: 72°C 45 sec *5. GO TO: 2 REP: 29 *6. T: 72°C 5min *7. HOLD: 10°C ResultsWe will have the results tomorrow
Culture for glycerol stocks and MiniPreps
Protocol :
Isolation of coloniesWe isolated colonies of several strains on a petri dish (streaked):
Culture : 37°C overnight |
 "
"